7OVG
 
 | |
7OAJ
 
 | | Crystal structure of pseudokinase CASK in complex with compound 7 | | Descriptor: | 1,2-ETHANEDIOL, 4-(cyclopentylamino)-2-[(3,4-dichlorophenyl)methylamino]-N-[3-(2-oxidanylidenepyrrolidin-1-yl)propyl]pyrimidine-5-carboxamide, Peripheral plasma membrane protein CASK | | Authors: | Chaikuad, A, Russ, N, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-19 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Design and Development of a Chemical Probe for Pseudokinase Ca 2+ /calmodulin-Dependent Ser/Thr Kinase.
J.Med.Chem., 64, 2021
|
|
7OAI
 
 | | Crystal structure of pseudokinase CASK in complex with PFE-PKIS12 | | Descriptor: | 1,2-ETHANEDIOL, 4-(Cyclopentylamino)-2-[(2,5-dichlorophenyl)methylamino]-N-[3-(2-oxo-1,3-oxazolidin-3-yl)propyl]pyrimidine-5-carboxamide, Peripheral plasma membrane protein CASK | | Authors: | Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-19 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and Development of a Chemical Probe for Pseudokinase Ca 2+ /calmodulin-Dependent Ser/Thr Kinase.
J.Med.Chem., 64, 2021
|
|
7OAK
 
 | | Crystal structure of pseudokinase CASK in complex with compound 26 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[2,5-bis(bromanyl)-4-methyl-phenyl]methylamino]-4-(cyclopentylamino)-N-[3-(2-oxidanylidene-1,3-oxazolidin-3-yl)propyl]pyrimidine-5-carboxamide, Peripheral plasma membrane protein CASK | | Authors: | Chaikuad, A, Russ, N, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-19 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Design and Development of a Chemical Probe for Pseudokinase Ca 2+ /calmodulin-Dependent Ser/Thr Kinase.
J.Med.Chem., 64, 2021
|
|
7OAL
 
 | | Crystal structure of pseudokinase CASK in complex with compound 25 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[2,5-bis(bromanyl)-4-methyl-phenyl]methylamino]-4-(cyclohexylamino)-N-[3-(2-oxidanylidene-1,3-oxazolidin-3-yl)propyl]pyrimidine-5-carboxamide, Peripheral plasma membrane protein CASK | | Authors: | Chaikuad, A, Russ, N, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-19 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Design and Development of a Chemical Probe for Pseudokinase Ca 2+ /calmodulin-Dependent Ser/Thr Kinase.
J.Med.Chem., 64, 2021
|
|
7OAM
 
 | | Kinase domain of MERTK in complex with compound 8 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[2,5-bis(fluoranyl)phenyl]methylamino]-4-(cyclopentylamino)-N-[3-(2-oxidanylidenepyrrolidin-1-yl)propyl]pyrimidine-5-carboxamide, Tyrosine-protein kinase Mer | | Authors: | Schroeder, M, Russ, N, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-19 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Design and Development of a Chemical Probe for Pseudokinase Ca 2+ /calmodulin-Dependent Ser/Thr Kinase.
J.Med.Chem., 64, 2021
|
|
4WXI
 
 | |
1MZG
 
 | | X-Ray Structure of SufE from E.coli Northeast Structural Genomics (NESG) Consortium Target ER30 | | Descriptor: | SufE Protein | | Authors: | Kuzin, A, Edstrom, W.C, Xiao, R, Acton, T.B, Rost, B, Tong, L, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-10-07 | | Release date: | 2003-01-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The SufE sulfur-acceptor protein contains a conserved core structure that mediates interdomain interactions in a variety of redox protein complexes
J.Mol.Biol., 344, 2004
|
|
8GBX
 
 | |
8GBY
 
 | | Crystal structure of PC39-50E, an anti-HIV broadly neutralizing antibody | | Descriptor: | GLYCEROL, PC39-50E Fab heavy chain, PC39-50E Fab light chain | | Authors: | Murrell, S, Omorodion, O, Wilson, I.A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Antigen pressure from two founder viruses induces multiple insertions at a single antibody position to generate broadly neutralizing HIV antibodies.
Plos Pathog., 19, 2023
|
|
8GBW
 
 | | Crystal structure of PC39-23D, an anti-HIV broadly neutralizing antibody | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PC39-23D Fab heavy chain, ... | | Authors: | Murrell, S, Omorodion, O, Wilson, I.A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-06-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Antigen pressure from two founder viruses induces multiple insertions at a single antibody position to generate broadly neutralizing HIV antibodies.
Plos Pathog., 19, 2023
|
|
8GBV
 
 | | Crystal structure of PC39-17A, an anti-HIV broadly neutralizing antibody | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, PC39-17A Fab heavy chain, ... | | Authors: | Murrell, S, Omorodion, O, Wilson, I.A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Antigen pressure from two founder viruses induces multiple insertions at a single antibody position to generate broadly neutralizing HIV antibodies.
Plos Pathog., 19, 2023
|
|
8GC0
 
 | | Crystal structure of PC39-50L, an anti-HIV broadly neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PC39-50L Fab heavy chain, PC39-50L Fab light chain | | Authors: | Murrell, S, Omorodion, O, Wilson, I.A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Antigen pressure from two founder viruses induces multiple insertions at a single antibody position to generate broadly neutralizing HIV antibodies.
Plos Pathog., 19, 2023
|
|
4LOH
 
 | | Crystal structure of hSTING(H232) in complex with c[G(2',5')pA(3',5')p] | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Gao, P, Patel, D.J. | | Deposit date: | 2013-07-12 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Function Analysis of STING Activation by c[G(2',5')pA(3',5')p] and Targeting by Antiviral DMXAA.
Cell(Cambridge,Mass.), 154, 2013
|
|
4LOJ
 
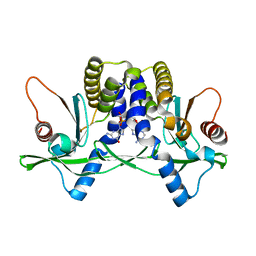 | | Crystal structure of mSting in complex with c[G(2',5')pA(3',5')p] | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Gao, P, Patel, D.J. | | Deposit date: | 2013-07-12 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure-Function Analysis of STING Activation by c[G(2',5')pA(3',5')p] and Targeting by Antiviral DMXAA.
Cell(Cambridge,Mass.), 154, 2013
|
|
8GC1
 
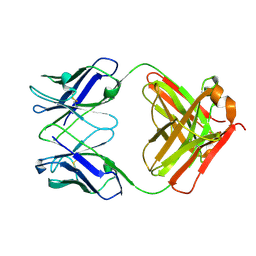 | |
3T22
 
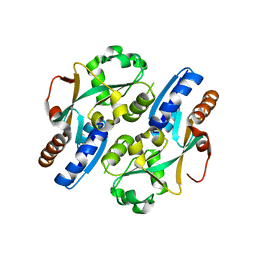 | |
8GBZ
 
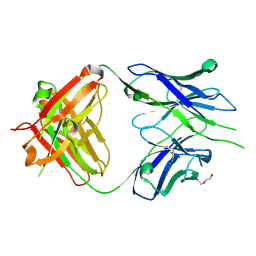 | | Crystal structure of PC39-55C, an anti-HIV broadly neutralizing antibody | | Descriptor: | DI(HYDROXYETHYL)ETHER, PC39-55C Fab heavy chain, PC39-55C Fab light chain | | Authors: | Murrell, S, Omorodion, O, Wilson, I.A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Antigen pressure from two founder viruses induces multiple insertions at a single antibody position to generate broadly neutralizing HIV antibodies.
Plos Pathog., 19, 2023
|
|
6TF7
 
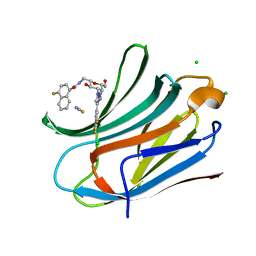 | | Human galectin-3c in complex with a galactose derivative | | Descriptor: | 4-fluoranyl-~{N}-[[(2~{S},3~{R},4~{R},5~{R},6~{R})-6-(hydroxymethyl)-3,5-bis(oxidanyl)-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxan-2-yl]methyl]naphthalene-1-carboxamide, CHLORIDE ION, Galectin-3, ... | | Authors: | Nilsson, U.J, Zetterberg, F, Hakansson, M, Logan, D.T. | | Deposit date: | 2019-11-13 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 3-Substituted 1-Naphthamidomethyl-C-galactosyls Interact with Two Unique Sub-sites for High-Affinity and High-Selectivity Inhibition of Galectin-3.
Molecules, 24, 2019
|
|
7U0P
 
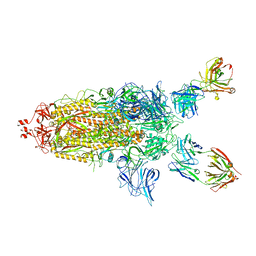 | | SARS-Cov2 S protein structure in complex with neutralizing monoclonal antibody 002-S21F2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Patel, A, Ortlund, E. | | Deposit date: | 2022-02-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structural insights for neutralization of Omicron variants BA.1, BA.2, BA.4, and BA.5 by a broadly neutralizing SARS-CoV-2 antibody.
Sci Adv, 8, 2022
|
|
1CZF
 
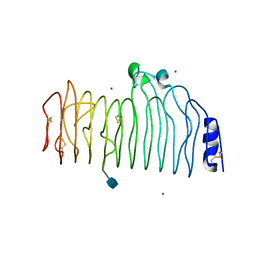 | | ENDO-POLYGALACTURONASE II FROM ASPERGILLUS NIGER | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, POLYGALACTURONASE II, ZINC ION | | Authors: | van Santen, Y, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1999-09-02 | | Release date: | 1999-10-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | 1.68-A crystal structure of endopolygalacturonase II from Aspergillus niger and identification of active site residues by site-directed mutagenesis.
J.Biol.Chem., 274, 1999
|
|
6TF6
 
 | | Human galectin-3c in complex with a galactose derivative | | Descriptor: | CHLORIDE ION, Galectin-3, ~{N}-[[(2~{S},3~{S},4~{R},5~{S},6~{R})-4-[[5,6-bis(fluoranyl)-2-oxidanylidene-chromen-3-yl]methoxy]-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]methyl]-4-fluoranyl-naphthalene-1-carboxamide | | Authors: | Nilsson, U.J, Zetterberg, F, Hakansson, M, Logan, D.T. | | Deposit date: | 2019-11-13 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 3-Substituted 1-Naphthamidomethyl-C-galactosyls Interact with Two Unique Sub-sites for High-Affinity and High-Selectivity Inhibition of Galectin-3.
Molecules, 24, 2019
|
|
4LOI
 
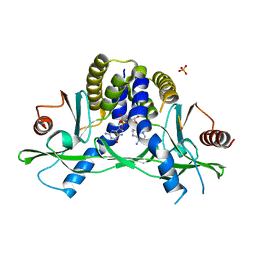 | | Crystal structure of hSTING(H232) in complex with c[G(2',5')pA(2',5')p] | | Descriptor: | 2-amino-9-[(1R,3R,6R,8R,9R,11S,14R,16R,17R,18R)-16-(6-amino-9H-purin-9-yl)-3,11,17,18-tetrahydroxy-3,11-dioxido-2,4,7,10,12,15-hexaoxa-3,11-diphosphatricyclo[12.2.1.1~6,9~]octadec-8-yl]-1,9-dihydro-6H-purin-6-one, PHOSPHATE ION, Stimulator of interferon genes protein | | Authors: | Gao, P, Patel, D.J. | | Deposit date: | 2013-07-12 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure-Function Analysis of STING Activation by c[G(2',5')pA(3',5')p] and Targeting by Antiviral DMXAA.
Cell(Cambridge,Mass.), 154, 2013
|
|
4LOK
 
 | | Crystal structure of mSting in complex with c[G(3',5')pA(3',5')p] | | Descriptor: | 2-amino-9-[(2R,3R,3aR,5S,7aS,9R,10R,10aR,12R,14aS)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, Stimulator of interferon genes protein | | Authors: | Gao, P, Patel, D.J. | | Deposit date: | 2013-07-12 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Function Analysis of STING Activation by c[G(2',5')pA(3',5')p] and Targeting by Antiviral DMXAA.
Cell(Cambridge,Mass.), 154, 2013
|
|
7TZ7
 
 | | PI3K alpha in complex with an inhibitor | | Descriptor: | (4S,5R)-3-[2'-amino-2-(morpholin-4-yl)-4'-(trifluoromethyl)[4,5'-bipyrimidin]-6-yl]-4-(hydroxymethyl)-5-methyl-1,3-oxazolidin-2-one, Isoform 3 of Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Knapp, M.S, Tang, J. | | Deposit date: | 2022-02-15 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Identification of NVP-CLR457 as an Orally Bioavailable Non-CNS-Penetrant pan-Class IA Phosphoinositol-3-Kinase Inhibitor.
J.Med.Chem., 65, 2022
|
|
