6ZDG
 
 | | Association of three complexes of largely structurally disordered Spike ectodomain with bound EY6A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EY6A heavy chain, EY6A light chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-06-14 | | Release date: | 2020-07-29 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZER
 
 | | Crystal structure of receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with EY6A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EY6A heavy chain, EY6A light chain, ... | | Authors: | Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-06-16 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5E06
 
 | |
5E05
 
 | |
8YRP
 
 | | SARS-CoV-2 Delta Spike in complex with JM-1A | | Descriptor: | JM-1A Heavy Chain, JM-1A Light Chain, Spike glycoprotein | | Authors: | Nguyen, V.H.T, Chen, X. | | Deposit date: | 2024-03-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
8YZ5
 
 | | SARS-CoV-2 Delta Spike in complex with Fab of JE-5C | | Descriptor: | Fab heavy chain of JE-5C, Fab light chain of JE-5C, Spike glycoprotein | | Authors: | Chen, X, Wu, Y.-M. | | Deposit date: | 2024-04-06 | | Release date: | 2024-06-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
8YRO
 
 | | SARS-CoV-2 Delta Spike in complex with JL-8C | | Descriptor: | JL-8C Heavy Chain, JL-8C Light Chain, Spike glycoprotein | | Authors: | Nguyen, V.H.T, Chen, X. | | Deposit date: | 2024-03-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
8YZ6
 
 | | SARS-CoV-2 Spike (BA.1) in complex with Fab of JH-8B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain of JH-8B, ... | | Authors: | Chen, X, Wu, Y.-M. | | Deposit date: | 2024-04-06 | | Release date: | 2024-06-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
6ZCZ
 
 | | Crystal structure of receptor binding domain of SARS-CoV-2 Spike glycoprotein in ternary complex with EY6A Fab and a nanobody. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, EY6A heavy chain, ... | | Authors: | Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-06-12 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5CEO
 
 | | DLK in complex with inhibitor 2-((6-(3,3-difluoropyrrolidin-1-yl)-4-(1-(oxetan-3-yl)piperidin-4-yl)pyridin-2-yl)amino)isonicotinonitrile | | Descriptor: | 2-[[6-[3,3-bis(fluoranyl)pyrrolidin-1-yl]-4-[1-(oxetan-3-yl)piperidin-4-yl]pyridin-2-yl]amino]pyridine-4-carbonitrile, Mitogen-activated protein kinase kinase kinase 12 | | Authors: | HARRIS, S.F, YIN, J. | | Deposit date: | 2015-07-07 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Scaffold-Hopping and Structure-Based Discovery of Potent, Selective, And Brain Penetrant N-(1H-Pyrazol-3-yl)pyridin-2-amine Inhibitors of Dual Leucine Zipper Kinase (DLK, MAP3K12).
J.Med.Chem., 58, 2015
|
|
5CIO
 
 | | Crystal structure of PqqF | | Descriptor: | ZINC ION, pyrroloquinoline quinone biosynthesis protein PqqF | | Authors: | Wei, Q, Xu, D, Ran, T, Wang, W. | | Deposit date: | 2015-07-13 | | Release date: | 2016-06-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure and Function of PqqF Protein in the Pyrroloquinoline Quinone Biosynthetic Pathway
J.Biol.Chem., 291, 2016
|
|
5CEN
 
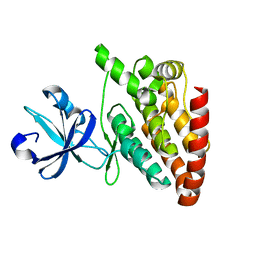 | | Crystal structure of DLK (kinase domain) | | Descriptor: | Mitogen-activated protein kinase kinase kinase 12 | | Authors: | HARRIS, S.F, YIN, J. | | Deposit date: | 2015-07-07 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Scaffold-Hopping and Structure-Based Discovery of Potent, Selective, And Brain Penetrant N-(1H-Pyrazol-3-yl)pyridin-2-amine Inhibitors of Dual Leucine Zipper Kinase (DLK, MAP3K12).
J.Med.Chem., 58, 2015
|
|
5E04
 
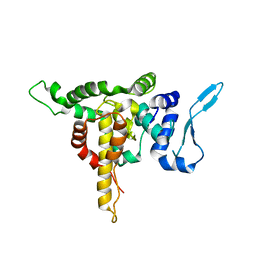 | | Crystal structure of Andes virus nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Wang, W.M, Lou, Z.Y. | | Deposit date: | 2015-09-28 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the Core Region of Hantavirus Nucleocapsid Protein Reveals the Mechanism for Ribonucleoprotein Complex Formation
J.Virol., 90, 2015
|
|
2LY0
 
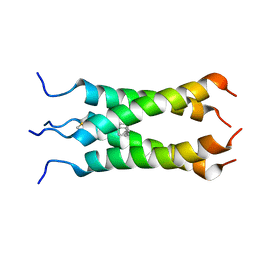 | | Solution NMR structure of the influenza A virus S31N mutant (19-49) in presence of drug M2WJ332 | | Descriptor: | (3S,5S,7S)-N-{[5-(thiophen-2-yl)-1,2-oxazol-3-yl]methyl}tricyclo[3.3.1.1~3,7~]decan-1-aminium, Membrane ion channel M2 | | Authors: | Wu, Y, Wang, J, DeGrado, W. | | Deposit date: | 2012-09-10 | | Release date: | 2013-01-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and inhibition of the drug-resistant S31N mutant of the M2 ion channel of influenza A virus.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2MUV
 
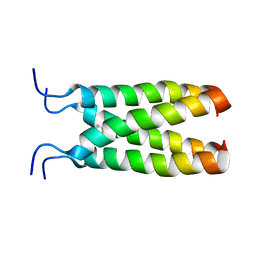 | | NOE-based model of the influenza A virus M2 (19-49) bound to drug 11 | | Descriptor: | (3s,5s,7s)-N-[(5-bromothiophen-2-yl)methyl]tricyclo[3.3.1.1~3,7~]decan-1-aminium, Matrix protein 2 | | Authors: | Wu, Y, Wang, J, DeGrado, W. | | Deposit date: | 2014-09-18 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Flipping in the Pore: Discovery of Dual Inhibitors That Bind in Different Orientations to the Wild-Type versus the Amantadine-Resistant S31N Mutant of the Influenza A Virus M2 Proton Channel.
J.Am.Chem.Soc., 136, 2014
|
|
5CEP
 
 | |
5CEQ
 
 | |
2MUW
 
 | | NOE-based model of the influenza A virus N31S mutant (19-49) bound to drug 11 | | Descriptor: | (3s,5s,7s)-N-[(5-bromothiophen-2-yl)methyl]tricyclo[3.3.1.1~3,7~]decan-1-aminium, Matrix protein 2 | | Authors: | Wu, Y, Wang, J, DeGrado, W. | | Deposit date: | 2014-09-18 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Flipping in the Pore: Discovery of Dual Inhibitors That Bind in Different Orientations to the Wild-Type versus the Amantadine-Resistant S31N Mutant of the Influenza A Virus M2 Proton Channel.
J.Am.Chem.Soc., 136, 2014
|
|
4PNU
 
 | | E. coli sliding clamp in complex with (R)-6-bromo-9-(2-((R)-1-carboxy-2-phenylethylamino)-2-oxoethyl)-2,3,4,9-tetrahydro-1H-carbazole-2-carboxylic acid | | Descriptor: | (2R)-6-bromo-9-(2-{[(1R)-1-carboxy-2-phenylethyl]amino}-2-oxoethyl)-2,3,4,9-tetrahydro-1H-carbazole-2-carboxylic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2014-02-21 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bacterial Sliding Clamp Inhibitors that Mimic the Sequential Binding Mechanism of Endogenous Linear Motifs.
J.Med.Chem., 58, 2015
|
|
4OVH
 
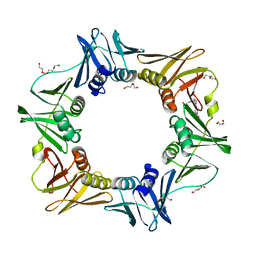 | | E. coli sliding clamp in complex with (R)-6-bromo-9-(2-(carboxymethylamino)-2-oxoethyl)-2,3,4,9-tetrahydro-1H-carbazole-2-carboxylic acid | | Descriptor: | (2R)-6-bromo-9-{2-[(carboxymethyl)amino]-2-oxoethyl}-2,3,4,9-tetrahydro-1H-carbazole-2-carboxylic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2014-02-21 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Bacterial Sliding Clamp Inhibitors that Mimic the Sequential Binding Mechanism of Endogenous Linear Motifs.
J.Med.Chem., 58, 2015
|
|
4PNW
 
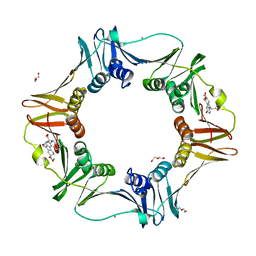 | | E. coli sliding clamp in complex with (R)-6-bromo-9-(2-((S)-1-carboxy-2-phenylethylamino)-2-oxoethyl)-2,3,4,9-tetrahydro-1H-carbazole-2-carboxylic acid | | Descriptor: | (2R)-6-bromo-9-(2-{[(1S)-1-carboxy-2-phenylethyl]amino}-2-oxoethyl)-2,3,4,9-tetrahydro-1H-carbazole-2-carboxylic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2014-02-21 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bacterial Sliding Clamp Inhibitors that Mimic the Sequential Binding Mechanism of Endogenous Linear Motifs.
J.Med.Chem., 58, 2015
|
|
4PNV
 
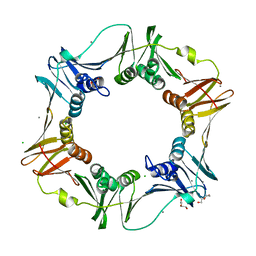 | |
7YCK
 
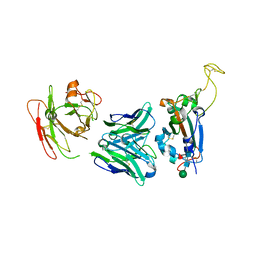 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with FP-12A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, FP-12A Fab heavy chain, FP-12A Fab light chain, ... | | Authors: | Nguyen, V.H.T, Chen, X. | | Deposit date: | 2022-07-01 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for a conserved neutralization epitope on the receptor-binding domain of SARS-CoV-2.
Nat Commun, 14, 2023
|
|
7YCL
 
 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with IS-9A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, IS-9A Fab heavy chain, IS-9A Fab light chain, ... | | Authors: | Mohapatra, A, Chen, X. | | Deposit date: | 2022-07-01 | | Release date: | 2023-02-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural basis for a conserved neutralization epitope on the receptor-binding domain of SARS-CoV-2.
Nat Commun, 14, 2023
|
|
7YCN
 
 | |
