7YQX
 
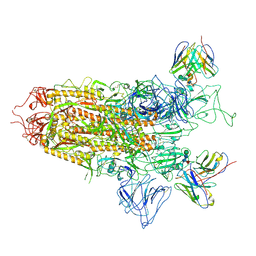 | | SARS-CoV-2 BA.2.75 S Trimer in complex with S309 (state1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 heavy chain, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YQZ
 
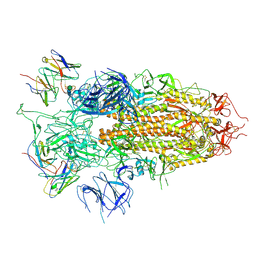 | | SARS-CoV-2 BA.2.75 S Trimer in complex with S309 (state3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S309 heavy chain, S309 light chain, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YQY
 
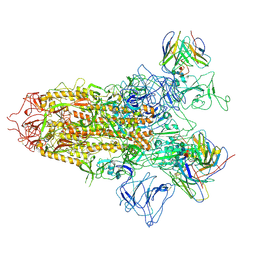 | | SARS-CoV-2 BA.2.75 S Trimer in complex with S309 (state2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 heavy chain, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
5Z0Q
 
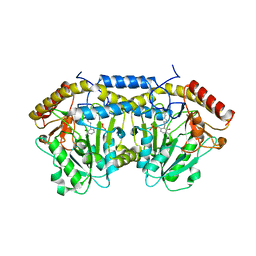 | | Crystal Structure of OvoB | | Descriptor: | Aminotransferase, class I and II, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Cai, Y.J, Huang, P, Wu, L, Zhou, J.H, Liu, P.H. | | Deposit date: | 2017-12-20 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | In Vitro Reconstitution of the Remaining Steps in Ovothiol A Biosynthesis: C-S Lyase and Methyltransferase Reactions.
Org. Lett., 20, 2018
|
|
7RND
 
 | | Crystal structure of caspase-3 with inhibitor Ac-VDPVD-CHO | | Descriptor: | Ac-VDPVD-CHO, Caspase-3 subunit p12, Caspase-3 subunit p17 | | Authors: | McCue, W, Finzel, B.C. | | Deposit date: | 2021-07-29 | | Release date: | 2022-01-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based Design and Biological Evaluation of Novel Caspase-2 Inhibitors Based on the Peptide AcVDVAD-CHO and the Caspase-2-Mediated Tau Cleavage Sequence YKPVD314.
Acs Pharmacol Transl Sci, 5, 2022
|
|
7RN8
 
 | | Crystal structure of caspase-3 with inhibitor Ac-VD(Orn)VD-CHO | | Descriptor: | Ac-VD(Orn)VD-CHO, Caspase-3 subunit p12, Caspase-3 subunit p17 | | Authors: | McCue, W, Finzel, B.C. | | Deposit date: | 2021-07-29 | | Release date: | 2022-01-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure-Based Design and Biological Evaluation of Novel Caspase-2 Inhibitors Based on the Peptide AcVDVAD-CHO and the Caspase-2-Mediated Tau Cleavage Sequence YKPVD314.
Acs Pharmacol Transl Sci, 5, 2022
|
|
7RNE
 
 | | Crystal structure of caspase-3 with inhibitor Ac-YKPVD-CHO | | Descriptor: | Ac-YKPVD-CHO, Caspase-3 subunit p12, Caspase-3 subunit p17 | | Authors: | McCue, W, Finzel, B.C. | | Deposit date: | 2021-07-29 | | Release date: | 2022-01-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structure-Based Design and Biological Evaluation of Novel Caspase-2 Inhibitors Based on the Peptide AcVDVAD-CHO and the Caspase-2-Mediated Tau Cleavage Sequence YKPVD314.
Acs Pharmacol Transl Sci, 5, 2022
|
|
7RN7
 
 | | Crystal structure of caspase-3 with inhibitor Ac-VD(Aly)VD-CHO | | Descriptor: | Ac-VD(Aly)VD-CHO, Caspase-3 subunit p12, Caspase-3 subunit p17 | | Authors: | McCue, W, Finzel, B.C. | | Deposit date: | 2021-07-29 | | Release date: | 2022-01-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Design and Biological Evaluation of Novel Caspase-2 Inhibitors Based on the Peptide AcVDVAD-CHO and the Caspase-2-Mediated Tau Cleavage Sequence YKPVD314.
Acs Pharmacol Transl Sci, 5, 2022
|
|
7RNB
 
 | | Crystal structure of caspase-3 with inhibitor Ac-VDRVD-CHO | | Descriptor: | Ac-VDRVD-CHO, Caspase-3 subunit p12, Caspase-3 subunit p17 | | Authors: | McCue, W, Finzel, B.C. | | Deposit date: | 2021-07-29 | | Release date: | 2022-01-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Based Design and Biological Evaluation of Novel Caspase-2 Inhibitors Based on the Peptide AcVDVAD-CHO and the Caspase-2-Mediated Tau Cleavage Sequence YKPVD314.
Acs Pharmacol Transl Sci, 5, 2022
|
|
7RN9
 
 | | Crystal structure of caspase-3 with inhibitor Ac-VDFVD-CHO | | Descriptor: | Ac-VDFVD-CHO, Caspase-3 subunit p12, Caspase-3 subunit p17 | | Authors: | McCue, W, Finzel, B.C. | | Deposit date: | 2021-07-29 | | Release date: | 2022-01-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure-Based Design and Biological Evaluation of Novel Caspase-2 Inhibitors Based on the Peptide AcVDVAD-CHO and the Caspase-2-Mediated Tau Cleavage Sequence YKPVD314.
Acs Pharmacol Transl Sci, 5, 2022
|
|
7RNF
 
 | | Crystal structure of caspase-3 with inhibitor Ac-VDKVD-CHO | | Descriptor: | Ac-VDKVD-CHO, Caspase-3 subunit p12, Caspase-3 subunit p17 | | Authors: | McCue, W, Finzel, B.C. | | Deposit date: | 2021-07-29 | | Release date: | 2022-01-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure-Based Design and Biological Evaluation of Novel Caspase-2 Inhibitors Based on the Peptide AcVDVAD-CHO and the Caspase-2-Mediated Tau Cleavage Sequence YKPVD314.
Acs Pharmacol Transl Sci, 5, 2022
|
|
7MCK
 
 | | Structure of CHK1 10-pt. mutant complex with LRRK2 inhibitor 18 | | Descriptor: | N-{5-[(3S)-3-(2-hydroxypropan-2-yl)pyrrolidin-1-yl]-2-(trifluoromethyl)pyridin-3-yl}-6-(1-methyl-1H-pyrazol-4-yl)pyridine-2-carboxamide, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R. | | Deposit date: | 2021-04-02 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Optimization of brain-penetrant picolinamide derived leucine-rich repeat kinase 2 (LRRK2) inhibitors.
Rsc Med Chem, 12, 2021
|
|
7L26
 
 | | HPK1 IN COMPLEX WITH COMPOUND 38 | | Descriptor: | 6-(2-fluoro-6-methylphenyl)-1-[4-(4-methylpiperazin-1-yl)phenyl]-1H-indazole-5-carbonitrile, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Lesburg, C.A. | | Deposit date: | 2020-12-16 | | Release date: | 2021-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of Potent Reverse Indazole Inhibitors for HPK1.
Acs Med.Chem.Lett., 12, 2021
|
|
7L25
 
 | | HPK1 IN COMPLEX WITH COMPOUND 18 | | Descriptor: | 1,2-ETHANEDIOL, 6-(2-fluoro-6-methoxyphenyl)-1-[6-(4-methylpiperazin-1-yl)pyridin-2-yl]-1H-pyrazolo[4,3-c]pyridine, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Lesburg, C.A. | | Deposit date: | 2020-12-16 | | Release date: | 2021-03-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification of Potent Reverse Indazole Inhibitors for HPK1.
Acs Med.Chem.Lett., 12, 2021
|
|
7L24
 
 | | HPK1 IN COMPLEX WITH COMPOUND 11 | | Descriptor: | 6-(2-fluoro-6-methoxyphenyl)-1-[4-(4-methylpiperazin-1-yl)phenyl]-1H-pyrazolo[4,3-c]pyridine, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Lesburg, C.A. | | Deposit date: | 2020-12-16 | | Release date: | 2021-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Identification of Potent Reverse Indazole Inhibitors for HPK1.
Acs Med.Chem.Lett., 12, 2021
|
|
5QTZ
 
 | | TGF-BETA RECEPTOR TYPE 1 KINASE DOMAIN (T204D) IN COMPLEX WITH 6-[1-(2,2-DIFLUOROETHYL)-4-(6-METHYLPYRIDIN-2-YL)-1H-IMIDAZOL-5-YL]IMIDAZO[1,2-A]PYRIDINE | | Descriptor: | 6-[1-(2,2-difluoroethyl)-4-(6-methylpyridin-2-yl)-1H-imidazol-5-yl]imidazo[1,2-a]pyridine, GLYCEROL, TGF-beta receptor type-1 | | Authors: | Sheriff, S. | | Deposit date: | 2019-11-19 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of BMS-986260, a Potent, Selective, and Orally Bioavailable TGF beta R1 Inhibitor as an Immuno-oncology Agent.
Acs Med.Chem.Lett., 11, 2020
|
|
5QU0
 
 | | TGF-BETA RECEPTOR TYPE 1 KINASE DOMAIN (T204D) IN COMPLEX WITH 6-[4-(3-CHLORO-4-FLUOROPHENYL)-1-(2-HYDROXYETHYL)-1H-IMIDAZOL-5-YL]IMIDAZO[1,2-B]PYRIDAZINE-3-CARBONITRILE | | Descriptor: | 6-[4-(3-chloro-4-fluorophenyl)-1-(2-hydroxyethyl)-1H-imidazol-5-yl]imidazo[1,2-b]pyridazine-3-carbonitrile, GLYCEROL, TGF-beta receptor type-1 | | Authors: | Sheriff, S. | | Deposit date: | 2019-11-19 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Discovery of BMS-986260, a Potent, Selective, and Orally Bioavailable TGF beta R1 Inhibitor as an Immuno-oncology Agent.
Acs Med.Chem.Lett., 11, 2020
|
|
6XLP
 
 | | Structure of the essential inner membrane lipopolysaccharide-PbgA complex | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-deoxy-3-O-[(1R,3R)-1,3-dihydroxytetradecyl]-2-{[(3R)-3-hydroxytetradecanoyl]amino}-1-O-phosphono-alpha-D-glucopyranose-(6-1)-[3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)]1,5-anhydro-2-deoxy-2-{[(1S,3R)-1-hydroxy-3-(pentanoyloxy)undecyl]amino}-4-O-phosphono-D-glucitol, ... | | Authors: | Payandeh, J, Clairefeuille, T. | | Deposit date: | 2020-06-29 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the essential inner membrane lipopolysaccharide-PbgA complex.
Nature, 584, 2020
|
|
6U2G
 
 | | BRAF-MEK complex with AMP-PCP bound to BRAF | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Liau, N.P.D, Wendorff, T, Hymowitz, S, Sudhamsu, J. | | Deposit date: | 2019-08-19 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.886 Å) | | Cite: | Negative regulation of RAF kinase activity by ATP is overcome by 14-3-3-induced dimerization.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7WRO
 
 | | Local structure of BD55-3372 and delta spike | | Descriptor: | 3372H, 3372L, Spike protein S1 | | Authors: | Liu, P.L. | | Deposit date: | 2022-01-27 | | Release date: | 2022-06-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
6U2H
 
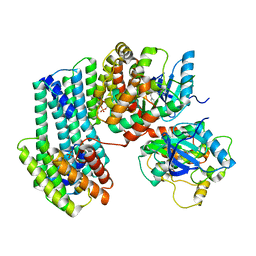 | | BRAF dimer bound to 14-3-3 | | Descriptor: | 14-3-3 protein zeta/delta, 2-{4-[(1E)-1-(hydroxyimino)-2,3-dihydro-1H-inden-5-yl]-3-(pyridin-4-yl)-1H-pyrazol-1-yl}ethanol, Serine/threonine-protein kinase B-raf | | Authors: | Liau, N.P.D, Hymowitz, S.G, Sudhamsu, J. | | Deposit date: | 2019-08-19 | | Release date: | 2019-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Negative regulation of RAF kinase activity by ATP is overcome by 14-3-3-induced dimerization.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3P9Y
 
 | |
7W06
 
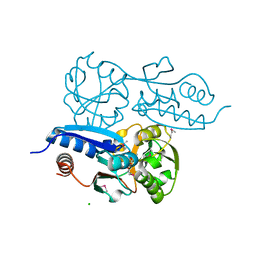 | | Itaconate inducible LysR-Type Transcriptional regulator (ITCR) in complex with itaconate (SeMet labeled), Space group C121. | | Descriptor: | 2-methylidenebutanedioic acid, CHLORIDE ION, SULFATE ION, ... | | Authors: | Sun, P.K, Wang, B, Wang, Z.X, Qi, S, Li, X.J. | | Deposit date: | 2021-11-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A genetically encoded fluorescent biosensor for detecting itaconate with subcellular resolution in living macrophages.
Nat Commun, 13, 2022
|
|
7W07
 
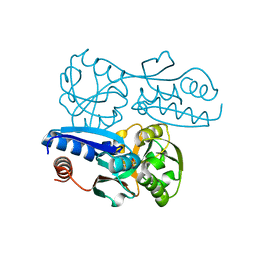 | | Itaconate inducible LysR-Type Transcriptional regulator (ITCR) in complex with itaconate, Space group C121. | | Descriptor: | 2-methylidenebutanedioic acid, SULFATE ION, Transcriptional regulator, ... | | Authors: | Sun, P.K, Wang, B, Li, X.J. | | Deposit date: | 2021-11-17 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | A genetically encoded fluorescent biosensor for detecting itaconate with subcellular resolution in living macrophages.
Nat Commun, 13, 2022
|
|
7W08
 
 | |
