1YEE
 
 | |
1YEC
 
 | |
1B8U
 
 | | MALATE DEHYDROGENASE FROM AQUASPIRILLUM ARCTICUM | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXALOACETATE ION, PROTEIN (MALATE DEHYDROGENASE) | | Authors: | Kim, S.Y, Hwang, K.Y, Kim, S.-H, Han, Y.S, Cho, Y. | | Deposit date: | 1999-02-02 | | Release date: | 1999-07-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for cold adaptation. Sequence, biochemical properties, and crystal structure of malate dehydrogenase from a psychrophile Aquaspirillium arcticum.
J.Biol.Chem., 274, 1999
|
|
1QVB
 
 | | CRYSTAL STRUCTURE OF THE BETA-GLYCOSIDASE FROM THE HYPERTHERMOPHILE THERMOSPHAERA AGGREGANS | | Descriptor: | BETA-GLYCOSIDASE | | Authors: | Chi, Y.-I, Martinez-Cruz, L.A, Swanson, R.V, Robertson, D.E, Kim, S.-H. | | Deposit date: | 1999-07-07 | | Release date: | 1999-07-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the beta-glycosidase from the hyperthermophile Thermosphaera aggregans: insights into its activity and thermostability.
FEBS Lett., 445, 1999
|
|
1B8P
 
 | | MALATE DEHYDROGENASE FROM AQUASPIRILLUM ARCTICUM | | Descriptor: | PROTEIN (MALATE DEHYDROGENASE) | | Authors: | Kim, S.Y, Hwang, K.Y, Kim, S.-H, Han, Y.S, Cho, Y. | | Deposit date: | 1999-02-02 | | Release date: | 1999-07-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for cold adaptation. Sequence, biochemical properties, and crystal structure of malate dehydrogenase from a psychrophile Aquaspirillium arcticum.
J.Biol.Chem., 274, 1999
|
|
1BCC
 
 | | CYTOCHROME BC1 COMPLEX FROM CHICKEN | | Descriptor: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, FE2/S2 (INORGANIC) CLUSTER, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.-I, Kim, K.K, Hung, L.-W, Crofts, A.R, Berry, E.A, Kim, S.-H. | | Deposit date: | 1998-03-23 | | Release date: | 1998-08-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Electron transfer by domain movement in cytochrome bc1.
Nature, 392, 1998
|
|
4LVH
 
 | | Insight into highly conserved H1 subtype-specific epitopes in influenza virus hemagglutinin | | Descriptor: | CALCIUM ION, Hemagglutinin, MONOCLONAL ANTIBODY H-CHAIN, ... | | Authors: | Kim, K.H, Cho, K.J, Kim, S, Seok, J.H, Lee, J.-H. | | Deposit date: | 2013-07-26 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insight into highly conserved h1 subtype-specific epitopes in influenza virus hemagglutinin
Plos One, 9, 2014
|
|
1KXU
 
 | | CYCLIN H, A POSITIVE REGULATORY SUBUNIT OF CDK ACTIVATING KINASE | | Descriptor: | CYCLIN H | | Authors: | Kim, K.K, Chamberin, H.M, Morgan, D.O, Kim, S.-H. | | Deposit date: | 1996-08-08 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three-dimensional structure of human cyclin H, a positive regulator of the CDK-activating kinase.
Nat.Struct.Biol., 3, 1996
|
|
1MOL
 
 | |
4NJD
 
 | | Structure of p21-activated kinase 4 with a novel inhibitor KY-04031 | | Descriptor: | N-(1H-indazol-5-yl)-N'-[2-(1H-indol-3-yl)ethyl]-6-methoxy-1,3,5-triazine-2,4-diamine, Serine/threonine-protein kinase PAK 4 | | Authors: | Park, S. | | Deposit date: | 2013-11-09 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and the structural basis of a novel p21-activated kinase 4 inhibitor.
Cancer Lett., 349, 2014
|
|
1LIH
 
 | | THREE-DIMENSIONAL STRUCTURES OF THE LIGAND-BINDING DOMAIN OF THE BACTERIAL ASPARTATE RECEPTOR WITH AND WITHOUT A LIGAND | | Descriptor: | 1,10-PHENANTHROLINE, ASPARTATE RECEPTOR | | Authors: | Kim, S.-H, Scott, W, Yeh, J.I, Prive, G.G, Milburn, M. | | Deposit date: | 1995-04-18 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand.
Science, 254, 1991
|
|
2EXM
 
 | | Human CDK2 in complex with isopentenyladenine | | Descriptor: | Cell division protein kinase 2, N-(3-METHYLBUT-2-EN-1-YL)-9H-PURIN-6-AMINE | | Authors: | Schulze-Gahmen, U. | | Deposit date: | 2005-11-08 | | Release date: | 2005-12-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Multiple modes of ligand recognition: crystal structures of cyclin-dependent protein kinase 2 in complex with ATP and two inhibitors, olomoucine and isopentenyladenine.
Proteins, 22, 1995
|
|
5DH5
 
 | | PDE10 complexed with N-[(1-methylpyrazol-4-yl)methyl]-5-[[(1S,2S)-2-(2-pyridyl)cyclopropyl]methoxy]pyrazolo[1,5-a]pyrimidin-7-amine | | Descriptor: | MAGNESIUM ION, N-[(1-methyl-1H-pyrazol-4-yl)methyl]-5-{[(1S,2S)-2-(pyridin-2-yl)cyclopropyl]methoxy}pyrazolo[1,5-a]pyrimidin-7-amine, ZINC ION, ... | | Authors: | Yan, Y. | | Deposit date: | 2015-08-29 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of pyrazolopyrimidine phosphodiesterase 10A inhibitors for the treatment of schizophrenia.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5DH4
 
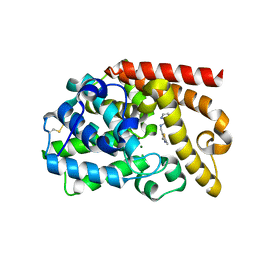 | | PDE10 complexed with 5-chloro-N-[(2,4-dimethylthiazol-5-yl)methyl]pyrazolo[1,5-a]pyrimidin-7-amine | | Descriptor: | 3-(1-hydroxy-2-methylpropan-2-yl)-5-phenyl-3,5-dihydro-1H-imidazo[4,5-c][1,8]naphthyridine-2,4-dione, 5-chloro-N-[(2,4-dimethyl-1,3-thiazol-5-yl)methyl]pyrazolo[1,5-a]pyrimidin-7-amine, MAGNESIUM ION, ... | | Authors: | Yan, Y. | | Deposit date: | 2015-08-29 | | Release date: | 2016-01-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of pyrazolopyrimidine phosphodiesterase 10A inhibitors for the treatment of schizophrenia.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
2FVO
 
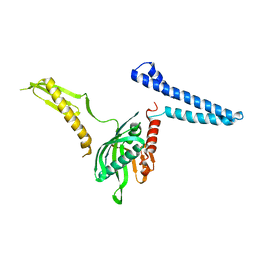 | | Docking of the modified RF1 X-ray structure into the Low Resolution Cryo-EM map of E.coli 70S Ribosome bound with RF1 | | Descriptor: | Peptide chain release factor 1 | | Authors: | Rawat, U, Gao, H, Zavialov, A, Gursky, R, Ehrenberg, M, Frank, J. | | Deposit date: | 2006-01-31 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12.8 Å) | | Cite: | Interactions of the Release Factor RF1 with the Ribosome as Revealed by Cryo-EM.
J.Mol.Biol., 357, 2006
|
|
4Q21
 
 | |
1LAG
 
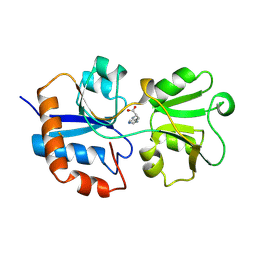 | | STRUCTURAL BASES FOR MULTIPLE LIGAND SPECIFICITY OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN | | Descriptor: | HISTIDINE, LYSINE, ARGININE, ... | | Authors: | Kim, S.-H, Oh, B.-H. | | Deposit date: | 1993-10-06 | | Release date: | 1995-07-10 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein.
J.Biol.Chem., 269, 1994
|
|
1LAF
 
 | | STRUCTURAL BASES FOR MULTIPLE LIGAND SPECIFICITY OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN | | Descriptor: | ARGININE, LYSINE, ORNITHINE-BINDING PROTEIN | | Authors: | Kim, S.-H, Oh, B.-H. | | Deposit date: | 1993-10-06 | | Release date: | 1995-07-10 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein.
J.Biol.Chem., 269, 1994
|
|
1LAH
 
 | | STRUCTURAL BASES FOR MULTIPLE LIGAND SPECIFICITY OF THE PERIPLASMIC LYSINE-, ARGININE-, ORNITHINE-BINDING PROTEIN | | Descriptor: | L-ornithine, LYSINE, ARGININE, ... | | Authors: | Kim, S.-H, Oh, B.-H. | | Deposit date: | 1993-10-06 | | Release date: | 1995-07-10 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein.
J.Biol.Chem., 269, 1994
|
|
3C4E
 
 | | Pim-1 Kinase Domain in Complex with 3-aminophenyl-7-azaindole | | Descriptor: | IMIDAZOLE, N-phenyl-1H-pyrrolo[2,3-b]pyridin-3-amine, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Zhang, K.Y.J, Wang, W. | | Deposit date: | 2008-01-29 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3C4F
 
 | |
3C4C
 
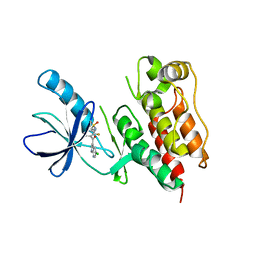 | | B-Raf Kinase in Complex with PLX4720 | | Descriptor: | B-Raf proto-oncogene serine/threonine-protein kinase, N-{3-[(5-chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)carbonyl]-2,4-difluorophenyl}propane-1-sulfonamide | | Authors: | Zhang, K.Y.J, Wang, W. | | Deposit date: | 2008-01-29 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1LST
 
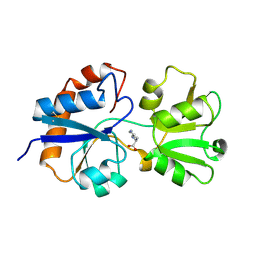 | |
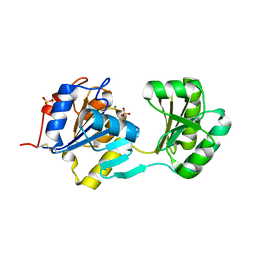
1NF2
 
 | |
1B0U
 
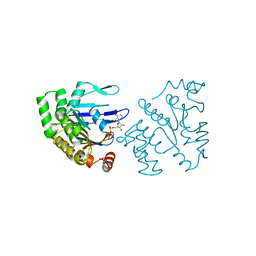 | | ATP-BINDING SUBUNIT OF THE HISTIDINE PERMEASE FROM SALMONELLA TYPHIMURIUM | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, HISTIDINE PERMEASE | | Authors: | Hung, L.-W, Wang, I.X, Nikaido, K, Liu, P.-Q, Ames, G.F.-L, Kim, S.-H. | | Deposit date: | 1998-11-12 | | Release date: | 1999-11-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the ATP-binding subunit of an ABC transporter.
Nature, 396, 1998
|
|
