6LLG
 
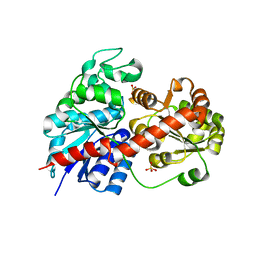 | | Crystal Structure of Fagopyrum esculentum M UGT708C1 | | Descriptor: | BENZAMIDINE, SULFATE ION, UDP-glycosyltransferase 708C1 | | Authors: | Wang, X, Liu, M. | | Deposit date: | 2019-12-23 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of theC-Glycosyltransferase UGT708C1 from Buckwheat Provide Insights into the Mechanism ofC-Glycosylation.
Plant Cell, 32, 2020
|
|
6LLZ
 
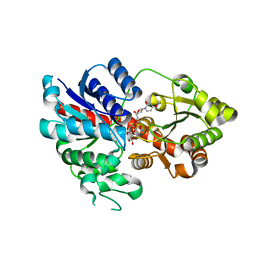 | |
1UP4
 
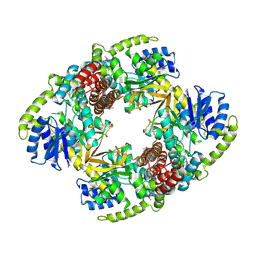 | |
8E3N
 
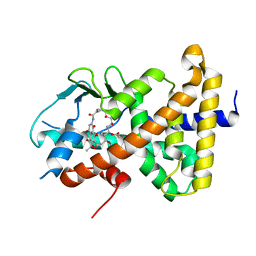 | | Crystal structure of pregnane X receptor ligand binding domain complexed with rifamycin S | | Descriptor: | Nuclear receptor subfamily 1 group I member 2, Rifamycin S | | Authors: | Huber, A.D, Poudel, S, Seetharaman, J, Miller, D.J, Lin, W, Chen, T. | | Deposit date: | 2022-08-17 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-guided approach to modulate small molecule binding to a promiscuous ligand-activated protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5H6I
 
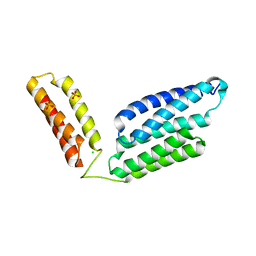 | | Crystal Structure of GBS CAMP Factor | | Descriptor: | CHLORIDE ION, Protein B, SULFATE ION | | Authors: | Jin, T.C, Brefo-Mensah, E.K. | | Deposit date: | 2016-11-13 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of theStreptococcus agalactiaeCAMP factor provides insights into its membrane-permeabilizing activity.
J.Biol.Chem., 293, 2018
|
|
7E3J
 
 | |
7E0B
 
 | |
4DOU
 
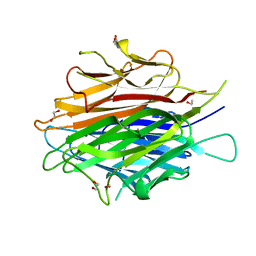 | | Crystal Structure of a Single-chain Trimer of Human Adiponectin Globular Domain | | Descriptor: | 1,2-ETHANEDIOL, Adiponectin, CALCIUM ION, ... | | Authors: | Min, X, Walker, N.P, Wang, Z. | | Deposit date: | 2012-02-10 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a single-chain trimer of human adiponectin globular domain.
Febs Lett., 586, 2012
|
|
8IPX
 
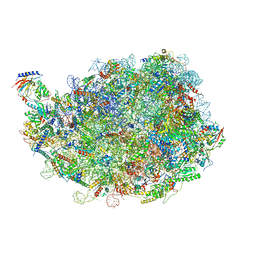 | |
8IPY
 
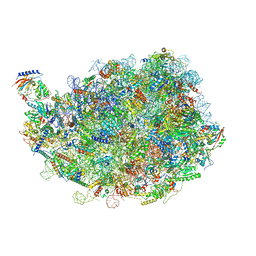 | |
8INK
 
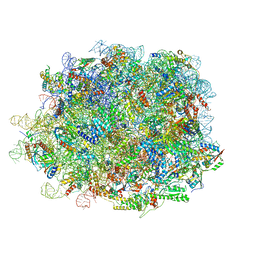 | | human nuclear pre-60S ribosomal particle - State D | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Zhang, Y, Gao, N. | | Deposit date: | 2023-03-10 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Visualizing the nucleoplasmic maturation of human pre-60S ribosomal particles.
Cell Res., 33, 2023
|
|
1UP7
 
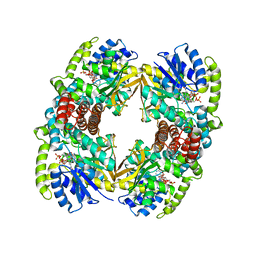 | | Structure of the 6-phospho-beta glucosidase from Thermotoga maritima at 2.4 Angstrom resolution in the tetragonal form with NAD and glucose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, 6-PHOSPHO-BETA-GLUCOSIDASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Varrot, A, Yip, V.L, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-09-29 | | Release date: | 2004-11-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Nad+ and Metal-Ion Dependent Hydrolysis by Family 4 Glycosidases: Structural Insight Into Specificity for Phospho-Beta-D-Glucosides
J.Mol.Biol., 346, 2005
|
|
5H5N
 
 | |
1B2D
 
 | |
4Z3C
 
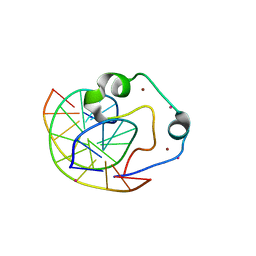 | | Zinc finger region of human TET3 in complex with CpG DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*CP*GP*TP*TP*GP*GP*C)-3'), Methylcytosine dioxygenase, UNKNOWN ATOM OR ION, ... | | Authors: | Liu, K, Xu, C, Tempel, W, Dong, A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-31 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | DNA Sequence Recognition of Human CXXC Domains and Their Structural Determinants.
Structure, 2017
|
|
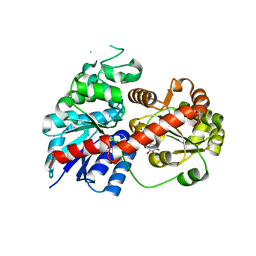
6LLW
 
 | |
5H3X
 
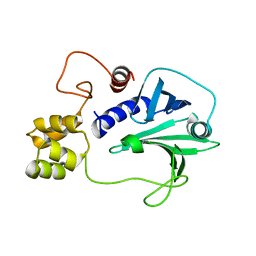 | |
6MIP
 
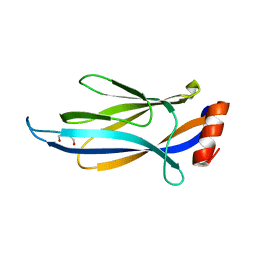 | |
4CU4
 
 | | FhuA from E. coli in complex with the lasso peptide microcin J25 (MccJ25) | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 3-HYDROXY-TETRADECANOIC ACID, DIPHOSPHATE, ... | | Authors: | Mathavan, I, Rebuffat, S, Beis, K. | | Deposit date: | 2014-03-17 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Hijacking Siderophore Receptors by Antimicrobial Lasso Peptides.
Nat.Chem.Biol., 10, 2014
|
|
7YHN
 
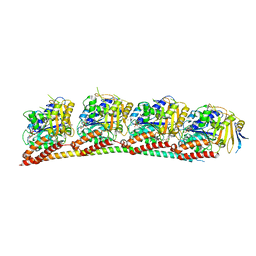 | | ANTI-TUMOR AGENT Y48 IN COMPLEX WITH TUBULIN | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-methyl-3-[(4-methylphenyl)sulfonylamino]-~{N}-[(6-methylpyridin-3-yl)methyl]benzamide, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Du, T, Ji, M, Hou, Z, Lin, S, Zhang, J, Wu, D, Zhang, K, Lu, D, Xu, H, Chen, X. | | Deposit date: | 2022-07-14 | | Release date: | 2023-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Optimization of Benzamide Derivatives as Potent and Orally Active Tubulin Inhibitors Targeting the Colchicine Binding Site.
J.Med.Chem., 65, 2022
|
|
3N7X
 
 | |
7ZE0
 
 | | Solution structure of the PulM C-terminal domain from Klebsiella oxytoca | | Descriptor: | Type II secretion system protein M | | Authors: | Lopez-Castilla, A, Bardiaux, B, Nilges, M, Francetic, O, Izadi-Pruneyre, N. | | Deposit date: | 2022-03-30 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamic association of an assembly platform subcomplex of the bacterial type II secretion system.
Structure, 31, 2023
|
|
8IJK
 
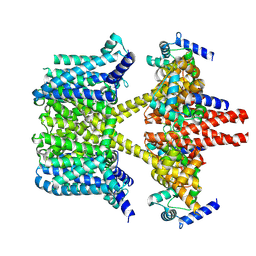 | | human KCNQ2-CaM-Ebio1 complex in the presence of PIP2 | | Descriptor: | Calmodulin-1, N-(1,2-dihydroacenaphthylen-5-yl)-4-fluoranyl-benzamide, Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Ma, D, Guo, J. | | Deposit date: | 2023-02-27 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A small-molecule activation mechanism that directly opens the KCNQ2 channel.
Nat.Chem.Biol., 20, 2024
|
|
7VLN
 
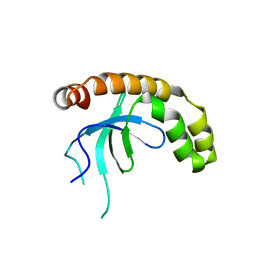 | | NSD2-PWWP1 domain bound with an imidazol-5-yl benzonitrile compound | | Descriptor: | 4-[5-[4-(aminomethyl)-2,6-dimethoxy-phenyl]-3-methyl-imidazol-4-yl]benzenecarbonitrile, Histone-lysine N-methyltransferase NSD2 | | Authors: | Cao, D.Y, Li, Y.L, Li, J, Xiong, B. | | Deposit date: | 2021-10-05 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structure-Based Discovery of a Series of NSD2-PWWP1 Inhibitors.
J.Med.Chem., 65, 2022
|
|
3FDR
 
 | | Crystal structure of TDRD2 | | Descriptor: | Tudor and KH domain-containing protein | | Authors: | Amaya, M.F, Adams, M.A, Guo, Y, Li, Y, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-26 | | Release date: | 2009-01-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
