8GH6
 
 | |
7VT1
 
 | |
7E3X
 
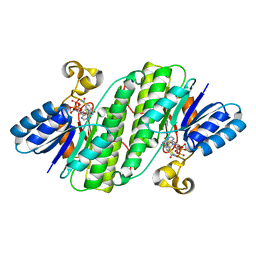 | | Crystal structure of SDR family NAD(P)-dependent oxidoreductase from exiguobacterium | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase | | Authors: | Chen, L, Tang, J, Yuan, S, Zhang, F, Chen, S. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure-guided evolution of a ketoreductase forefficient and stereoselective bioreduction of bulkyalpha-aminobeta-keto esters
Catalysis Science And Technology, 2021
|
|
9ERY
 
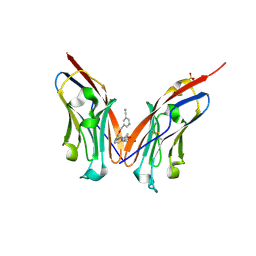 | | Co-crystal strucutre of PD-L1 with low molecular weight inhibitor | | Descriptor: | 5-[[5-[[2-[bis(fluoranyl)methyl]-3-(2,3-dihydro-1,4-benzodioxin-6-yl)phenyl]methoxy]-2-[(2-hydroxyethylamino)methyl]phenoxy]methyl]pyridine-3-carbonitrile, Programmed cell death 1 ligand 1, SULFATE ION | | Authors: | Plewka, J, Magiera-Mularz, K, Zhang, W. | | Deposit date: | 2024-03-25 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design, synthesis, and evaluation of antitumor activity of 2-arylmethoxy-4-(2-fluoromethyl-biphenyl-3-ylmethoxy) benzylamine derivatives as PD-1/PD-l1 inhibitors.
Eur.J.Med.Chem., 276, 2024
|
|
5JDR
 
 | | Structure of PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Zhou, A, Wei, H. | | Deposit date: | 2016-04-17 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of a novel PD-L1 nanobody for immune checkpoint blockade.
Cell Discov, 3, 2017
|
|
5JDS
 
 | |
3PH5
 
 | |
3PH6
 
 | |
8E7M
 
 | |
6RQM
 
 | |
9C9Q
 
 | |
9C9R
 
 | |
9CBS
 
 | |
6DNE
 
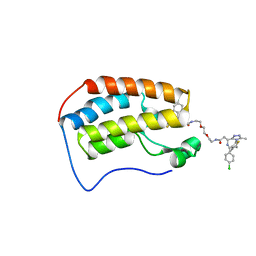 | | Crystal structure of human Bromodomain-containing protein 4 (BRD4) bromodomain with MS660 | | Descriptor: | Bromodomain-containing protein 4, N,N'-[ethane-1,2-diylbis(oxyethane-2,1-diyl)]bis{2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]acetamide} | | Authors: | Ren, C, Zhou, M.M. | | Deposit date: | 2018-06-06 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.958 Å) | | Cite: | Spatially constrained tandem bromodomain inhibition bolsters sustained repression of BRD4 transcriptional activity for TNBC cell growth.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DJC
 
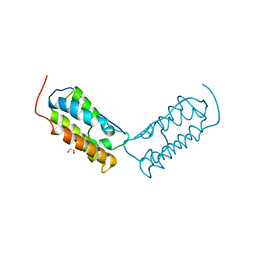 | | Crystal structure of human Bromodomain-containing protein 4 (BRD4) bromodomain with MS645 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N,N'-(decane-1,10-diyl)bis{2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]acetamide} | | Authors: | Ren, C, Zhou, M.M. | | Deposit date: | 2018-05-25 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Spatially constrained tandem bromodomain inhibition bolsters sustained repression of BRD4 transcriptional activity for TNBC cell growth.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7RN0
 
 | |
7RN1
 
 | |
7LTY
 
 | | Bruton's tyrosine kinase in complex with compound 23 | | Descriptor: | DIMETHYL SULFOXIDE, Isoform BTK-C of Tyrosine-protein kinase BTK, ~{N}-[(5~{R})-2-[2-[(1-methylpyrazol-4-yl)amino]pyrimidin-4-yl]-6,7,8,9-tetrahydro-5~{H}-benzo[7]annulen-5-yl]-3-propan-2-yloxy-azetidine-1-carboxamide | | Authors: | Metrick, C.M, Marcotte, D.J. | | Deposit date: | 2021-02-20 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery and Preclinical Characterization of BIIB091, a Reversible, Selective BTK Inhibitor for the Treatment of Multiple Sclerosis.
J.Med.Chem., 65, 2022
|
|
7LTZ
 
 | | Bruton's tyrosine kinase in complex with compound 51 | | Descriptor: | 1-~{tert}-butyl-~{N}-[(5~{R})-8-[2-[(1-methylpyrazol-4-yl)amino]pyrimidin-4-yl]-2-(oxetan-3-yl)-1,3,4,5-tetrahydro-2-benzazepin-5-yl]-1,2,3-triazole-4-carboxamide, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACETATE ION, ... | | Authors: | Metrick, C.M, Marcotte, D.J. | | Deposit date: | 2021-02-20 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Discovery and Preclinical Characterization of BIIB091, a Reversible, Selective BTK Inhibitor for the Treatment of Multiple Sclerosis.
J.Med.Chem., 65, 2022
|
|
5WY0
 
 | | Crystal structure of the methyltranferase domain of human HEN1 in complex with AdoMet | | Descriptor: | S-ADENOSYLMETHIONINE, Small RNA 2'-O-methyltransferase | | Authors: | Peng, L, Ma, J.B, Wu, L.G, Huang, Y. | | Deposit date: | 2017-01-10 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Identification of substrates of the small RNA methyltransferase Hen1 in mouse spermatogonial stem cells and analysis of its methyl-transfer domain
J. Biol. Chem., 293, 2018
|
|
5X2H
 
 | | Crystal structure of Campylobacter jejuni Cas9 in complex with sgRNA and target DNA (AGAAACA PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, Non-target DNA strand, ... | | Authors: | Yamada, M, Watanabe, Y, Hirano, H, Nakane, T, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2017-01-31 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Minimal Cas9 from Campylobacter jejuni Reveals the Molecular Diversity in the CRISPR-Cas9 Systems
Mol. Cell, 65, 2017
|
|
5X2G
 
 | | Crystal structure of Campylobacter jejuni Cas9 in complex with sgRNA and target DNA (AGAAACC PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, Non-target DNA strand, ... | | Authors: | Yamada, M, Watanabe, Y, Hirano, H, Nakane, T, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2017-01-31 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Minimal Cas9 from Campylobacter jejuni Reveals the Molecular Diversity in the CRISPR-Cas9 Systems
Mol. Cell, 65, 2017
|
|
5CHE
 
 | |
6UDN
 
 | | Crystal Structure of a Self-Assembling DNA Scaffold Containing TA Sticky Ends and Rhombohedral Symmetry | | Descriptor: | ARSENIC, DNA (5'-D(*TP*AP*CP*TP*GP*AP*CP*TP*CP*AP*TP*GP*CP*TP*CP*AP*TP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*AP*TP*CP*AP*GP*AP*TP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2019-09-19 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | A Self-Assembled Rhombohedral DNA Crystal Scaffold with Tunable Cavity Sizes and High-Resolution Structural Detail.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6U40
 
 | | Crystal Structure of a Self-Assembling DNA Crystal Scaffold with Rhombohedral Symmetry | | Descriptor: | CACODYLATE ION, DNA (5'-D(*CP*AP*CP*TP*GP*AP*CP*TP*CP*AP*TP*GP*CP*TP*CP*AP*TP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*AP*GP*AP*TP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2019-08-22 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | A Self-Assembled Rhombohedral DNA Crystal Scaffold with Tunable Cavity Sizes and High-Resolution Structural Detail.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
