6W52
 
 | | Prefusion RSV F bound by neutralizing antibody RSB1 | | Descriptor: | Fusion glycoprotein F0, Fusion glycoprotein F1 fused with Fibritin trimerization domain, RSB1 Fab Heavy Chain, ... | | Authors: | Harshbarger, W, Chandramouli, S, Malito, M. | | Deposit date: | 2020-03-12 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.74 Å) | | Cite: | Convergent structural features of respiratory syncytial virus neutralizing antibodies and plasticity of the site V epitope on prefusion F.
Plos Pathog., 16, 2020
|
|
6CRM
 
 | |
6DCE
 
 | | X-ray structure of FIP200 claw domain | | Descriptor: | RB1-inducible coiled-coil protein 1, SULFATE ION | | Authors: | Su, M.-Y, Hurley, J.H. | | Deposit date: | 2018-05-05 | | Release date: | 2019-03-06 | | Last modified: | 2019-05-01 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | FIP200 Claw Domain Binding to p62 Promotes Autophagosome Formation at Ubiquitin Condensates.
Mol. Cell, 74, 2019
|
|
4R2K
 
 | |
4R2J
 
 | |
4R2L
 
 | |
4R2M
 
 | |
1Y4H
 
 | | Wild type staphopain-staphostatin complex | | Descriptor: | CHLORIDE ION, SULFATE ION, cysteine protease, ... | | Authors: | Filipek, R, Potempa, J, Bochtler, M. | | Deposit date: | 2004-11-30 | | Release date: | 2005-01-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A comparison of staphostatin B with standard mechanism serine protease inhibitors.
J.Biol.Chem., 280, 2005
|
|
1RHB
 
 | | WATER DEPENDENT DOMAIN MOTION AND FLEXIBILITY IN RIBONUCLEASE A AND THE INVARIANT FEATURES IN ITS HYDRATION SHELL. AN X-RAY STUDY OF TWO LOW HUMIDITY CRYSTAL FORMS OF THE ENZYME | | Descriptor: | RIBONUCLEASE A | | Authors: | Radha Kishan, K.V, Chandra, N.R, Sudarsanakumar, C, Suguna, K, Vijayan, M. | | Deposit date: | 1994-11-13 | | Release date: | 1995-02-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Water-dependent domain motion and flexibility in ribonuclease A and the invariant features in its hydration shell. An X-ray study of two low-humidity crystal forms of the enzyme.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1RHA
 
 | | WATER DEPENDENT DOMAIN MOTION AND FLEXIBILITY IN RIBONUCLEASE A AND THE INVARIANT FEATURES IN ITS HYDRATION SHELL. AN X-RAY STUDY OF TWO LOW HUMIDITY CRYSTAL FORMS OF THE ENZYME | | Descriptor: | RIBONUCLEASE A | | Authors: | Radha Kishan, K.V, Chandra, N.R, Sudarsanakumar, C, Suguna, K, Vijayan, M. | | Deposit date: | 1994-11-13 | | Release date: | 1995-02-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Water-dependent domain motion and flexibility in ribonuclease A and the invariant features in its hydration shell. An X-ray study of two low-humidity crystal forms of the enzyme.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1XEK
 
 | |
1XEI
 
 | |
1XEJ
 
 | |
3KM4
 
 | | Optimization of Orally Bioavailable Alkyl Amine Renin Inhibitors | | Descriptor: | (3R)-3-[(1S)-4-(acetylamino)-1-(3-chlorophenyl)-1-hydroxybutyl]-N-{(1S)-2-cyclohexyl-1-[(methylamino)methyl]ethyl}piperidine-1-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, Z, McKeever, B.M. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimization of orally bioavailable alkyl amine renin inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4J3L
 
 | | Tankyrase 2 in complex with 3-chloro-N-(2-methoxyethyl)-4-(4-methyl-2-oxo-1,2-dihydroquinolin-7-yl)benzamide | | Descriptor: | 3-chloro-N-(2-methoxyethyl)-4-(4-methyl-2-oxo-1,2-dihydroquinolin-7-yl)benzamide, SULFATE ION, Tankyrase-2, ... | | Authors: | Jansson, A.E, Larsson, E.A, Nordlund, P.L. | | Deposit date: | 2013-02-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Fragment-based ligand design of novel potent inhibitors of tankyrases.
J.Med.Chem., 56, 2013
|
|
4J1Z
 
 | |
4J21
 
 | | Tankyrase 2 in complex with 7-(4-amino-2-chlorophenyl)-4-methylquinolin-2(1H)-one | | Descriptor: | 7-(4-amino-2-chlorophenyl)-4-methylquinolin-2(1H)-one, SULFATE ION, Tankyrase-2, ... | | Authors: | Jansson, A.E, Larsson, E.A, Nordlund, P.L. | | Deposit date: | 2013-02-04 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Fragment-based ligand design of novel potent inhibitors of tankyrases.
J.Med.Chem., 56, 2013
|
|
3NBN
 
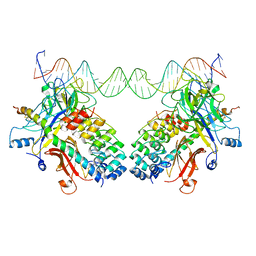 | |
4J22
 
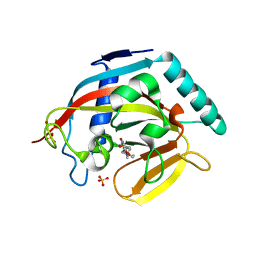 | | Tankyrase 2 in complex with 3-chloro-4-(4-methyl-2-oxo-1,2-dihydroquinolin-7-yl)-N-[2-(morpholin-4-yl)ethyl]benzamide | | Descriptor: | 3-chloro-4-(4-methyl-2-oxo-1,2-dihydroquinolin-7-yl)-N-[2-(morpholin-4-yl)ethyl]benzamide, SULFATE ION, Tankyrase-2, ... | | Authors: | Jansson, A.E, Larsson, E.A, Nordlund, P.L. | | Deposit date: | 2013-02-04 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Fragment-based ligand design of novel potent inhibitors of tankyrases.
J.Med.Chem., 56, 2013
|
|
4IUE
 
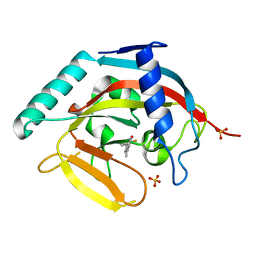 | | Tankyrase in complex with 7-(2-fluorophenyl)-4-methyl-1,2-dihydroquinolin-2-one | | Descriptor: | 7-(2-fluorophenyl)-4-methylquinolin-2(1H)-one, SULFATE ION, Tankyrase-2, ... | | Authors: | Jansson, A.E, Larsson, E.A, Nordlund, P.L. | | Deposit date: | 2013-01-21 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Fragment-based ligand design of novel potent inhibitors of tankyrases.
J.Med.Chem., 56, 2013
|
|
4J3M
 
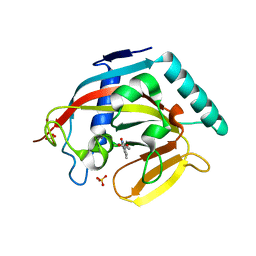 | | Tankyrase 2 in complex with 3-chloro-4-(4-methyl-2-oxo-1,2-dihydroquinolin-7-yl)benzoic acid | | Descriptor: | 3-chloro-4-(4-methyl-2-oxo-1,2-dihydroquinolin-7-yl)benzoic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Jansson, A.E, Larsson, E.A, Nordlund, P.L. | | Deposit date: | 2013-02-06 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-based ligand design of novel potent inhibitors of tankyrases.
J.Med.Chem., 56, 2013
|
|
3TFP
 
 | | Crystal Structure of Dehydrosqualene Synthase (CrtM) from S. aureus Complexed with BPH-1162 | | Descriptor: | 2-({2-chloro-6-[(2,4-dichlorophenyl)sulfanyl]benzyl}carbamoyl)benzoic acid, Dehydrosqualene synthase, MAGNESIUM ION | | Authors: | Lin, F.-Y, Liu, Y.-L, Zhang, Y, Oldfield, E. | | Deposit date: | 2011-08-16 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dual dehydrosqualene/squalene synthase inhibitors: leads for innate immune system-based therapeutics.
Chemmedchem, 7, 2012
|
|
3TFV
 
 | | Crystal structure of dehydrosqualene synthase (crtm) from s. aureus complexed with bph-1154 | | Descriptor: | 5-bromo-2-{[3-(octyloxy)benzyl]sulfanyl}benzoic acid, Dehydrosqualene synthase, MAGNESIUM ION | | Authors: | Lin, F.-Y, Zhang, Y, Liu, Y.-L, Oldfield, E. | | Deposit date: | 2011-08-16 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Dual dehydrosqualene/squalene synthase inhibitors: leads for innate immune system-based therapeutics.
Chemmedchem, 7, 2012
|
|
3TFN
 
 | |
2LZJ
 
 | | Refined solution structure and dynamics of First Catalytic Cysteine Half-domain from mouse E1 enzyme | | Descriptor: | Ubiquitin-like modifier-activating enzyme 1 | | Authors: | Jaremko, M, Jaremko, L, Nowakowski, M, Szczepanowski, R.H, Filipek, R, Wojciechowski, M, Bochtler, M, Ejchart, A. | | Deposit date: | 2012-10-03 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structural studies of the first catalytic half-domain of ubiquitin activating enzyme.
J.Struct.Biol., 185, 2014
|
|
