7XLJ
 
 | | The crystal structure of ORE1(ANAC092) NAC domain | | Descriptor: | NAC domain-containing protein 92 | | Authors: | Chun, I.S, Kim, M.S. | | Deposit date: | 2022-04-21 | | Release date: | 2023-03-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis of DNA binding by the NAC transcription factor ORE1, a master regulator of plant senescence.
Plant Commun., 4, 2023
|
|
7XP3
 
 | | DNA complex form of ORESARA1(ANAC092) NAC Domain | | Descriptor: | DNA (5'-D(*AP*GP*TP*TP*AP*CP*GP*TP*AP*CP*GP*GP*CP*AP*CP*AP*CP*GP*TP*AP*AP*C)-3'), DNA (5'-D(*TP*GP*TP*TP*AP*CP*GP*TP*GP*TP*GP*CP*CP*GP*TP*AP*CP*GP*TP*AP*AP*C)-3'), DNA (5'-D(P*CP*AP*CP*AP*CP*GP*TP*AP*AP*C)-3'), ... | | Authors: | Chun, I.S, Kim, M.S. | | Deposit date: | 2022-05-03 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural basis of DNA binding by the NAC transcription factor ORE1, a master regulator of plant senescence.
Plant Commun., 4, 2023
|
|
2NYI
 
 | | Crystal Structure of an Unknown Protein from Galdieria sulphuraria | | Descriptor: | unknown protein | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, McCoy, J.G, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-11-20 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of tandem ACT domain-containing protein ACTP from Galdieria sulphuraria.
Proteins, 80, 2012
|
|
1DU3
 
 | | Crystal structure of TRAIL-SDR5 | | Descriptor: | DEATH RECEPTOR 5, TNF-RELATED APOPTOSIS INDUCING LIGAND, ZINC ION | | Authors: | Cha, S.-S, Sung, B.-J, Oh, B.-H. | | Deposit date: | 2000-01-14 | | Release date: | 2000-09-27 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of TRAIL-DR5 complex identifies a critical role of the unique frame insertion in conferring recognition specificity
J.Biol.Chem., 275, 2000
|
|
2A5E
 
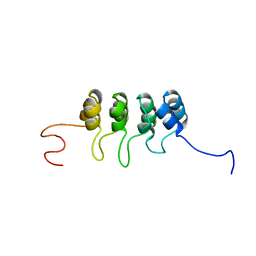 | | SOLUTION NMR STRUCTURE OF TUMOR SUPPRESSOR P16INK4A, RESTRAINED MINIMIZED MEAN STRUCTURE | | Descriptor: | TUMOR SUPPRESSOR P16INK4A | | Authors: | Byeon, I.-J.L, Li, J, Ericson, K, Selby, T.L, Tevelev, A, Kim, H.-J, O'Maille, P, Tsai, M.-D. | | Deposit date: | 1998-02-13 | | Release date: | 1999-08-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor p16INK4A: determination of solution structure and analyses of its interaction with cyclin-dependent kinase 4.
Mol.Cell, 1, 1998
|
|
2GM3
 
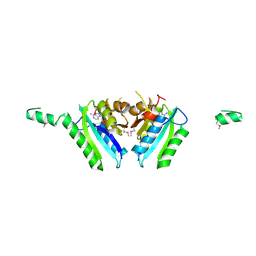 | | Crystal Structure of an Universal Stress Protein Family Protein from Arabidopsis Thaliana At3g01520 with AMP Bound | | Descriptor: | ADENOSINE MONOPHOSPHATE, unknown protein | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-04-05 | | Release date: | 2006-04-18 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.461 Å) | | Cite: | Crystal structure of the protein At3g01520, a eukaryotic universal stress protein-like protein from arabidopsis thaliana in complex with AMP.
Proteins, 83, 2015
|
|
5U9M
 
 | |
5YCN
 
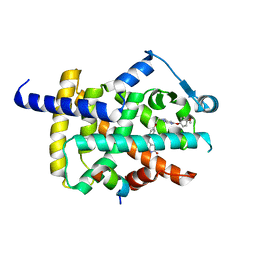 | | Human PPARgamma ligand binding domain complexed with Lobeglitazone | | Descriptor: | (5S)-5-[[4-[2-[[6-(4-methoxyphenoxy)pyrimidin-4-yl]-methyl-amino]ethoxy]phenyl]methyl]-1,3-thiazolidine-2,4-dione, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Jang, J.Y, Han, B.W. | | Deposit date: | 2017-09-07 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for the Enhanced Anti-Diabetic Efficacy of Lobeglitazone on PPAR gamma.
Sci Rep, 8, 2018
|
|
5YCP
 
 | | Human PPARgamma ligand binding domain complexed with Rosiglitazone | | Descriptor: | 2,4-THIAZOLIDIINEDIONE, 5-[[4-[2-(METHYL-2-PYRIDINYLAMINO)ETHOXY]PHENYL]METHYL]-(9CL), Nuclear receptor coactivator 1, ... | | Authors: | Jang, J.Y, Han, B.W. | | Deposit date: | 2017-09-07 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Enhanced Anti-Diabetic Efficacy of Lobeglitazone on PPAR gamma.
Sci Rep, 8, 2018
|
|
6A0H
 
 | |
6A0F
 
 | |
6A0I
 
 | |
6A0E
 
 | |
6IJR
 
 | | Human PPARgamma ligand binding domain complexed with SB1495 | | Descriptor: | 16 mer peptide from Nuclear receptor coactivator 1, N-{[3-({[(1S,2R)-2-{[(2E)-2-cyano-4,4-dimethylpent-2-enoyl]amino}cyclopentyl]oxy}methyl)phenyl]methyl}-4-[(4-methylpiperazin-1-yl)methyl]benzamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Jang, J.Y, Han, B.W. | | Deposit date: | 2018-10-11 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis for the inhibitory effects of a novel reversible covalent ligand on PPAR gamma phosphorylation.
Sci Rep, 9, 2019
|
|
5CEH
 
 | | Structure of histone lysine demethylase KDM5A in complex with selective inhibitor | | Descriptor: | 7-oxo-5-phenyl-6-(propan-2-yl)-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonitrile, Lysine-specific demethylase 5A, NICKEL (II) ION, ... | | Authors: | Kiefer, J.R, Vinogradova, M. | | Deposit date: | 2015-07-06 | | Release date: | 2016-05-18 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | An inhibitor of KDM5 demethylases reduces survival of drug-tolerant cancer cells.
Nat.Chem.Biol., 12, 2016
|
|
6IY6
 
 | | Crystal structure of human cytosolic aspartyl-tRNA synthetase (DRS) in complex with glutathion-S transferase (GST) domains from Aminoacyl tRNA synthase complex-interacting multifunctional protein 2 (AIMP2) and glutamyl-prolyl-tRNA synthetase (EPRS) | | Descriptor: | Aminoacyl tRNA synthase complex-interacting multifunctional protein 2, Aspartate--tRNA ligase, cytoplasmic, ... | | Authors: | Park, S.H, Hahn, H, Han, B.W. | | Deposit date: | 2018-12-13 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The DRS-AIMP2-EPRS subcomplex acts as a pivot in the multi-tRNA synthetase complex.
Iucrj, 6, 2019
|
|
7M2U
 
 | | Nucleotide Excision Repair complex TFIIH Rad4-33 | | Descriptor: | CALCIUM ION, DNA repair helicase RAD25, DNA repair helicase RAD3, ... | | Authors: | van Eeuwen, T, Murakami, K. | | Deposit date: | 2021-03-17 | | Release date: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Cryo-EM structure of TFIIH/Rad4-Rad23-Rad33 in damaged DNA opening in nucleotide excision repair.
Nat Commun, 12, 2021
|
|
7MKA
 
 | | Structure of EC+EC (leading EC-focused) | | Descriptor: | DNA (40-MER), DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Murakami, K. | | Deposit date: | 2021-04-22 | | Release date: | 2022-04-27 | | Last modified: | 2023-05-17 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7MK9
 
 | | Complex structure of trailing EC of EC+EC (trailing EC-focused) | | Descriptor: | DNA (40-MER), DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Murakami, K. | | Deposit date: | 2021-04-22 | | Release date: | 2022-04-27 | | Last modified: | 2023-05-17 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
6JQ7
 
 | |
7MEI
 
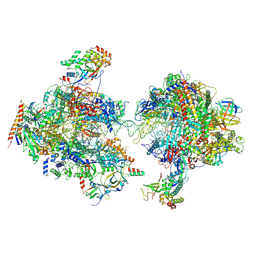 | | Composite structure of EC+EC | | Descriptor: | DNA (74-MER), DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Murakami, K. | | Deposit date: | 2021-04-06 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
6IJS
 
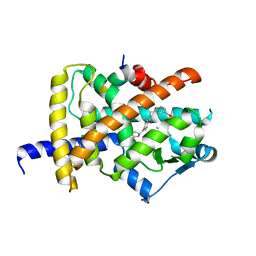 | | Human PPARgamma ligand binding domain complexed with SB1494 | | Descriptor: | 16-mer peptide from Nuclear receptor coactivator 1, N-{[3-({[(1R,2S)-2-{[(2E)-2-cyano-4,4-dimethylpent-2-enoyl]amino}cyclopentyl]oxy}methyl)phenyl]methyl}-4-[(4-methylpiperazin-1-yl)methyl]benzamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Jang, J.Y, Han, B.W. | | Deposit date: | 2018-10-11 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for the inhibitory effects of a novel reversible covalent ligand on PPAR gamma phosphorylation.
Sci Rep, 9, 2019
|
|
2K21
 
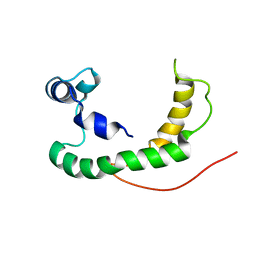 | | NMR structure of human KCNE1 in LMPG micelles at pH 6.0 and 40 degree C | | Descriptor: | Potassium voltage-gated channel subfamily E member | | Authors: | Kang, C, Tian, C, Sonnichsen, F.D, Smith, J.A, Meiler, J, George, A.L, Vanoye, C.G, Sanders, C.R, Kim, H. | | Deposit date: | 2008-03-19 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of KCNE1 and implications for how it modulates the KCNQ1 potassium channel.
Biochemistry, 47, 2008
|
|
2KDC
 
 | | NMR Solution Structure of E. coli diacylglycerol kinase (DAGK) in DPC micelles | | Descriptor: | Diacylglycerol kinase | | Authors: | Van Horn, W.D, Kim, H, Ellis, C.D, Hadziselimovic, A, Sulistijo, E.S, Karra, M.D, Tian, C, Sonnichsen, F.D, Sanders, C.R. | | Deposit date: | 2009-01-06 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution nuclear magnetic resonance structure of membrane-integral diacylglycerol kinase
Science, 324, 2009
|
|
3SJF
 
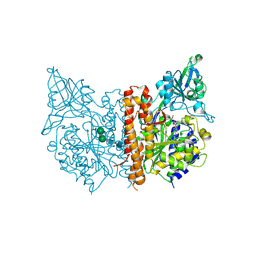 | | X-ray structure of human glutamate carboxypeptidase II in complex with a urea-based inhibitor (A25) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Plechanovova, A, Byun, Y, Alquicer, G, Skultetyova, L, Mlcochova, P, Nemcova, A, Kim, H, Navratil, M, Mease, R, Lubkowski, J, Pomper, M, Konvalinka, J, Rulisek, L, Barinka, C. | | Deposit date: | 2011-06-21 | | Release date: | 2011-10-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Novel Substrate-Based Inhibitors of Human Glutamate Carboxypeptidase II with Enhanced Lipophilicity.
J.Med.Chem., 54, 2011
|
|
