1K3N
 
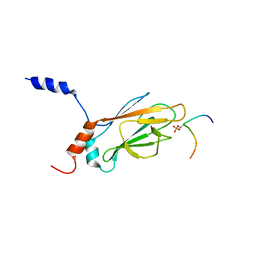 | | NMR Structure of the FHA1 Domain of Rad53 in Complex with a Rad9-derived Phosphothreonine (at T155) Peptide | | Descriptor: | DNA repair protein Rad9, Protein Kinase SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-03 | | Release date: | 2001-12-05 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
3C7O
 
 | | Crystal structure of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from Bacillus subtilis in complex with cellotetraose. | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase, FORMIC ACID, ... | | Authors: | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | Deposit date: | 2008-02-08 | | Release date: | 2008-11-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
1J4P
 
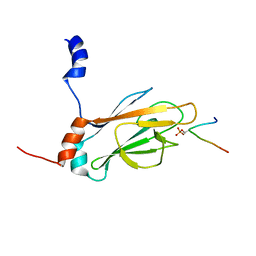 | | NMR STRUCTURE OF THE FHA1 DOMAIN OF RAD53 IN COMPLEX WITH A RAD9-DERIVED PHOSPHOTHREONINE (AT T155) PEPTIDE | | Descriptor: | DNA REPAIR PROTEIN RAD9, PROTEIN KINASE SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-22 | | Release date: | 2001-12-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
3C7F
 
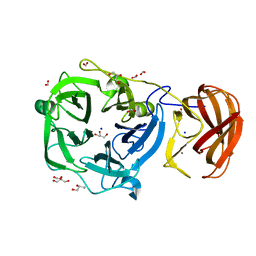 | | Crystal structure of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from bacillus subtilis in complex with xylotriose. | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase, FORMIC ACID, ... | | Authors: | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | Deposit date: | 2008-02-07 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
3C7H
 
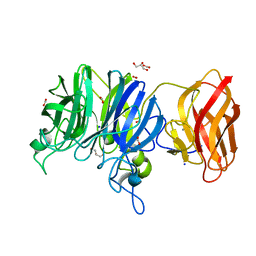 | | Crystal structure of glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from Bacillus subtilis in complex with AXOS-4-0.5. | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase, FORMIC ACID, ... | | Authors: | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | Deposit date: | 2008-02-07 | | Release date: | 2008-11-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
3C7G
 
 | | Crystal structure of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from Bacillus subtilis in complex with xylotetraose. | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase, GLYCEROL, ... | | Authors: | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | Deposit date: | 2008-02-07 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
3C7E
 
 | | Crystal structure of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from Bacillus subtilis. | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase, FORMIC ACID, ... | | Authors: | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | Deposit date: | 2008-02-07 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
1HLK
 
 | | METALLO-BETA-LACTAMASE FROM BACTEROIDES FRAGILIS IN COMPLEX WITH A TRICYCLIC INHIBITOR | | Descriptor: | 7,8-DIHYDROXY-1-METHOXY-3-METHYL-10-OXO-4,10-DIHYDRO-1H,3H-PYRANO[4,3-B]CHROMENE-9-CARBOXYLIC ACID, BETA-LACTAMASE, TYPE II, ... | | Authors: | Payne, D.J, Hueso-Rodriguez, J.A, Boyd, H, Concha, N.O, Janson, C.A, Gilpin, M, Bateson, J.H, Chever, C, Niconovich, N.L, Pearson, S, Rittenhouse, S, Tew, D, Diez, E, Perez, P, de la Fuente, J, Rees, M, Rivera-Sagredo, A. | | Deposit date: | 2000-12-01 | | Release date: | 2001-11-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a series of tricyclic natural products as potent broad spectrum inhibitors of metallo-beta-lactamases
Antimicrob.Agents Chemother., 46, 2002
|
|
1K3J
 
 | | Refined NMR Structure of the FHA1 Domain of Yeast Rad53 | | Descriptor: | Protein Kinase SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-03 | | Release date: | 2001-12-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
1KR3
 
 | | Crystal Structure of the Metallo beta-Lactamase from Bacteroides fragilis (CfiA) in Complex with the Tricyclic Inhibitor SB-236050. | | Descriptor: | 7,8-DIHYDROXY-1-METHOXY-3-METHYL-10-OXO-4,10-DIHYDRO-1H,3H-PYRANO[4,3-B]CHROMENE-9-CARBOXYLIC ACID, SODIUM ION, ZINC ION, ... | | Authors: | Payne, D.J, Hueso-Rodrguez, J.A, Boyd, H, Concha, N.O, Janson, C.A, Gilpin, M, Bateson, J.H, Cheever, C, Niconovich, N.L, Pearson, S, Rittenhouse, S, Tew, D, Dez, E, Prez, P, de la Fuente, J, Rees, M, Rivera-Sagredo, A. | | Deposit date: | 2002-01-08 | | Release date: | 2003-01-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a series of tricyclic natural products as potent broad-spectrum inhibitors of metallo-beta-lactamases
ANTIMICROB.AGENTS CHEMOTHER., 46, 2002
|
|
8J4I
 
 | | Unveiling the Role of Human Prohibitin 2 as a Mitochondrial Calcium Channel in Parkinson's Disease | | Descriptor: | Prohibitin-2 | | Authors: | Li, Y.Y, Fang, Y, Gao, Y.X, Ding, W, Yang, J, Liu, Y, Shen, B, Wang, J.F, Zhou, S. | | Deposit date: | 2023-04-20 | | Release date: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Human Prohibitin 2 is a Mitochondrial Ca2+-Selective Channel
To Be Published
|
|
1J4O
 
 | | REFINED NMR STRUCTURE OF THE FHA1 DOMAIN OF YEAST RAD53 | | Descriptor: | PROTEIN KINASE SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-03 | | Release date: | 2001-12-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
3ROF
 
 | | Crystal Structure of the S. aureus Protein Tyrosine Phosphatase PtpA | | Descriptor: | Expression tag cleaved from protein-tyrosine-phosphatase ptpA, Low molecular weight protein-tyrosine-phosphatase ptpA, PHOSPHATE ION | | Authors: | Grundner, C, Chou, S, Engel, K. | | Deposit date: | 2011-04-25 | | Release date: | 2011-09-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Structure and substrate recognition of the Staphylococcus aureus protein tyrosine phosphatase PtpA.
J.Mol.Biol., 413, 2011
|
|
5UYO
 
 | | Solution NMR structure of the de novo mini protein HEEH_rd4_0097 | | Descriptor: | HEEH_rd4_0097 | | Authors: | Lemak, A, Rocklin, G.J, Houliston, S, Carter, L, Chidyausiku, T.M, Baker, D, Arrowsmith, C.H. | | Deposit date: | 2017-02-24 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Global analysis of protein folding using massively parallel design, synthesis, and testing.
Science, 357, 2017
|
|
3M7R
 
 | | Crystal structure of VDR H305Q mutant | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, Vitamin D3 receptor | | Authors: | Rochel, N, Hourai, S, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2010-03-17 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of hereditary vitamin D-resistant rickets--associated mutant H305Q of vitamin D nuclear receptor bound to its natural ligand
J.Steroid Biochem.Mol.Biol., 121, 2010
|
|
4P3X
 
 | | Structure of the Fe4S4 quinolinate synthase NadA from Thermotoga maritima | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, IRON/SULFUR CLUSTER, Quinolinate synthase A, ... | | Authors: | Cherrier, M.V, Chan, A, Darnault, C, Reichmann, D, Amara, P, Ollagnier de Choudens, S, Fontecilla-Camps, J.C. | | Deposit date: | 2014-03-10 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of Fe4S4 quinolinate synthase unravels an enzymatic dehydration mechanism that uses tyrosine and a hydrolase-type triad.
J.Am.Chem.Soc., 136, 2014
|
|
2LFE
 
 | | Solution NMR structure of N-terminal domain of human E3 ubiquitin-protein ligase HECW2, Northeast structural genomics consortium (NESG) target ht6306A | | Descriptor: | E3 ubiquitin-protein ligase HECW2 | | Authors: | Lemak, A, Yee, A, Houliston, S, Garcia, M, Chitayat, S, Dhe-Paganon, S, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-29 | | Release date: | 2011-07-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of N-terminal domain of E3 ligase HECW2
To be Published
|
|
4LYC
 
 | | Cd ions within a lysoyzme single crystal | | Descriptor: | CADMIUM ION, Lysozyme C | | Authors: | Wei, H, House, S, Wu, J, Zhang, J, Wang, Z, He, Y, Gao, Y.-G, Robinson, H, Li, W, Zuo, J.-M, Robertson, I.M, Lu, Y. | | Deposit date: | 2013-07-30 | | Release date: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Enhanced and tunable fluorescent quantum dots within a single crystal of protein
TO BE PUBLISHED
|
|
4LYB
 
 | | CdS within a lysoyzme single crystal | | Descriptor: | CADMIUM ION, Lysozyme C | | Authors: | Wei, H, House, S, Wu, J, Zhang, J, Wang, Z, He, Y, Gao, Y.-G, Robinson, H, Li, W, Zuo, J.-M, Robertson, I.M, Lu, Y. | | Deposit date: | 2013-07-30 | | Release date: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Enhanced and tunable fluorescent quantum dots within a single crystal of protein
TO BE PUBLISHED
|
|
1XCC
 
 | | 1-Cys peroxidoxin from Plasmodium Yoelli | | Descriptor: | 1-Cys peroxiredoxin | | Authors: | Vedadi, M, Sharma, S, Houston, S, Lew, J, Wasney, G, Amani, M, Xu, X, Bray, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Hui, R, Bochkarev, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2004-09-01 | | Release date: | 2004-11-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
1Z8U
 
 | | Crystal structure of oxidized alpha hemoglobin bound to AHSP | | Descriptor: | Alpha-hemoglobin stabilizing protein, Hemoglobin alpha chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Feng, L, Zhou, S, Gu, L, Gell, D.A, Mackay, J.P, Weiss, M.J, Gow, A.J, Shi, Y. | | Deposit date: | 2005-03-31 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of oxidized alpha-haemoglobin bound to AHSP reveals a protective mechanism for haem.
Nature, 435, 2005
|
|
1Y01
 
 | | Crystal structure of AHSP bound to Fe(II) alpha-hemoglobin | | Descriptor: | 6-[(CYCLOHEXYLACETYL)(2-HYDROXYETHYL)AMINO]-6-DEOXY-D-XYLO-HEXITOL, Alpha-hemoglobin stabilizing protein, Hemoglobin alpha chain, ... | | Authors: | Feng, L, Gell, D.A, Zhou, S, Gu, L, Gow, A.J, Weiss, M.J, Mackay, J.P, Shi, Y. | | Deposit date: | 2004-11-14 | | Release date: | 2004-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular mechanism of AHSP-mediated stabilization of alpha-hemoglobin.
Cell(Cambridge,Mass.), 119, 2004
|
|
4KT3
 
 | | Structure of a type VI secretion system effector-immunity complex from Pseudomonas protegens | | Descriptor: | Putative lipoprotein, Uncharacterized protein | | Authors: | Whitney, J.C, Chou, S, Gardiner, T.E, Mougous, J.D. | | Deposit date: | 2013-05-19 | | Release date: | 2013-07-31 | | Last modified: | 2013-10-09 | | Method: | X-RAY DIFFRACTION (1.4362 Å) | | Cite: | Identification, Structure, and Function of a Novel Type VI Secretion Peptidoglycan Glycoside Hydrolase Effector-Immunity Pair.
J.Biol.Chem., 288, 2013
|
|
1XZY
 
 | | Solution structure of the P30-trans form of Alpha Hemoglobin Stabilizing Protein (AHSP) | | Descriptor: | Alpha-hemoglobin stabilizing protein | | Authors: | Gell, D.A, Feng, L, Zhou, S, Kong, Y, Lee, C, Weiss, M.J, Shi, Y, Mackay, J.P. | | Deposit date: | 2004-11-12 | | Release date: | 2004-12-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Molecular mechanism of AHSP-mediated stabilization of alpha-hemoglobin
Cell(Cambridge,Mass.), 119, 2004
|
|
1TXJ
 
 | | Crystal structure of translationally controlled tumour-associated protein (TCTP) from Plasmodium knowlesi | | Descriptor: | translationally controlled tumour-associated protein (TCTP) from Plasmodium knowlesi, PKN_PFE0545c | | Authors: | Walker, J.R, Vedadi, M, Sharma, S, Houston, S, Lew, J, Amani, M, Wasney, G, Skarina, T, Bray, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2004-07-05 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
