8D1B
 
 | |
7REP
 
 | |
7REO
 
 | |
3RZ2
 
 | | Crystal of Prl-1 complexed with peptide | | Descriptor: | Prl-1 (PTP4A1), Protein tyrosine phosphatase type IVA 1 | | Authors: | Zhang, Z.-Y, Liu, D, Bai, Y. | | Deposit date: | 2011-05-11 | | Release date: | 2011-10-26 | | Last modified: | 2014-10-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | PRL-1 protein promotes ERK1/2 and RhoA protein activation through a non-canonical interaction with the Src homology 3 domain of p115 Rho GTPase-activating protein.
J.Biol.Chem., 286, 2011
|
|
7VU8
 
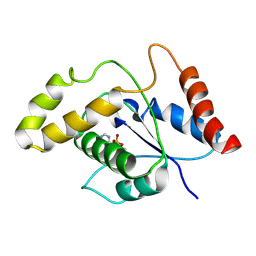 | | L7-Tir domain with bound ligand | | Descriptor: | 2',3'- cyclic AMP, Flax rust resistance protein | | Authors: | Tan, Y, Xu, C, Yu, D, Song, W, Wu, B, Schulze-Lefert, P, Chai, J. | | Deposit date: | 2021-11-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | TIR domains of plant immune receptors are 2',3'-cAMP/cGMP synthetases mediating cell death.
Cell, 185, 2022
|
|
6AF0
 
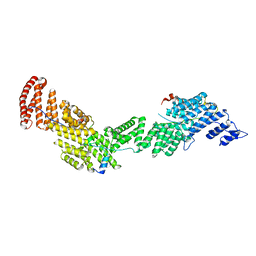 | | Structure of Ctr9, Paf1 and Cdc73 ternary complex from Myceliophthora thermophila | | Descriptor: | Cdc73 protein, Ctr9 protein, Paf1 protein | | Authors: | Wang, Z, Deng, P, Zhou, Y. | | Deposit date: | 2018-08-07 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Transcriptional elongation factor Paf1 core complex adopts a spirally wrapped solenoidal topology.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6A0P
 
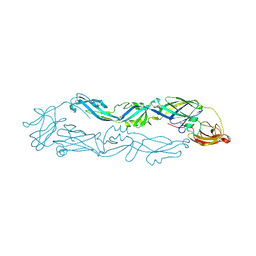 | | Crystal structure of Usutu virus envelope protein in the pre-fusion state | | Descriptor: | Envelope protein | | Authors: | Lu, G, Chen, Z, Ye, F, Lin, S, Yang, F, Cheng, Y. | | Deposit date: | 2018-06-06 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Usutu virus envelope protein in the pre-fusion state
Virol. J., 15, 2018
|
|
6IP1
 
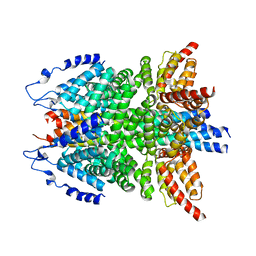 | | alpha-SNAP-SNARE subcomplex in the whole 20S complex | | Descriptor: | Alpha-soluble NSF attachment protein, Synaptosomal-associated protein 25, Syntaxin-1A, ... | | Authors: | Huang, X, Sun, S, Wang, X, Fan, F, Zhou, Q, Sui, S.F. | | Deposit date: | 2018-11-01 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanistic insights into the SNARE complex disassembly.
Sci Adv, 5, 2019
|
|
6IP2
 
 | | NSF-D1D2 part in the whole 20S complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Vesicle-fusing ATPase | | Authors: | Huang, X, Sun, S, Wang, X, Fan, F, Zhou, Q, Sui, S.F. | | Deposit date: | 2018-11-01 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanistic insights into the SNARE complex disassembly.
Sci Adv, 5, 2019
|
|
6IG9
 
 | |
5Z38
 
 | | Crystal structure of CsrA bound to CesT | | Descriptor: | CesT protein, Truncated-CsrA, wild type CsrA | | Authors: | Ye, F, Yang, F, Yu, R, Lin, X, Qi, J, Chen, Z, Gao, G.F, Lu, G. | | Deposit date: | 2018-01-05 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Molecular basis of binding between the global post-transcriptional regulator CsrA and the T3SS chaperone CesT
Nat Commun, 9, 2018
|
|
5F95
 
 | | Crystal structure of GSK3b in complex with Compound 18: 2-[(cyclopropylcarbonyl)amino]-N-(4-phenylpyridin-3-yl)pyridine-4-carboxamide | | Descriptor: | 2-[(cyclopropylcarbonyl)amino]-N-(4-phenylpyridin-3-yl)pyridine-4-carboxamide, Glycogen synthase kinase-3 beta | | Authors: | Lewis, H.A, Kish, K, Luo, G, Dubowchick, G.M. | | Deposit date: | 2015-12-09 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.525 Å) | | Cite: | Discovery of Isonicotinamides as Highly Selective, Brain Penetrable, and Orally Active Glycogen Synthase Kinase-3 Inhibitors.
J.Med.Chem., 59, 2016
|
|
5F94
 
 | | Crystal structure of GSK3b in complex with Compound 15: 2-[(cyclopropylcarbonyl)amino]-N-(4-methoxypyridin-3-yl)pyridine-4-carboxamide | | Descriptor: | 2-[(cyclopropylcarbonyl)amino]-N-(4-methoxypyridin-3-yl)pyridine-4-carboxamide, Glycogen synthase kinase-3 beta | | Authors: | Lewis, H.A, Kish, K, Luo, G, Dubowchick, G.M. | | Deposit date: | 2015-12-09 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery of Isonicotinamides as Highly Selective, Brain Penetrable, and Orally Active Glycogen Synthase Kinase-3 Inhibitors.
J.Med.Chem., 59, 2016
|
|
8GRR
 
 | | Complex of FMDV A/WH/CHA/09 and bovine neutralizing scFv antibody W125 | | Descriptor: | A/WH/CHA/09 VP1, A/WH/CHA/09 VP2, A/WH/CHA/09 VP3, ... | | Authors: | He, Y, Kun, L. | | Deposit date: | 2022-09-02 | | Release date: | 2023-10-11 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Conserved antigen structures and antibody-driven variations on foot-and-mouth disease virus serotype A revealed by bovine neutralizing monoclonal antibodies.
Plos Pathog., 19, 2023
|
|
8GSP
 
 | | Complex of FMDV A/WH/CHA/09 and bovine neutralizing scFv antibody W2 | | Descriptor: | A/WH/CHA/09 VP1, A/WH/CHA/09 VP2, A/WH/CHA/09 VP3, ... | | Authors: | He, Y, Li, K. | | Deposit date: | 2022-09-06 | | Release date: | 2023-10-11 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Conserved antigen structures and antibody-driven variations on foot-and-mouth disease virus serotype A revealed by bovine neutralizing monoclonal antibodies.
Plos Pathog., 19, 2023
|
|
8GYB
 
 | |
8GYA
 
 | |
8GY4
 
 | | Crystal structure of Alongshan virus methyltransferase | | Descriptor: | Methyltransferase | | Authors: | Chen, H, Lin, S, Lu, G.W. | | Deposit date: | 2022-09-21 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional basis of low-affinity SAM/SAH-binding in the conserved MTase of the multi-segmented Alongshan virus distantly related to canonical unsegmented flaviviruses.
Plos Pathog., 19, 2023
|
|
8GY9
 
 | |
8H5U
 
 | | Crystal structure of SARS-CoV-2 spike receptor-binding domain in complex with neutralizing nanobody Nb-021 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody Nb-021, ... | | Authors: | Yang, J, Lin, S, Lu, G.W. | | Deposit date: | 2022-10-13 | | Release date: | 2023-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Development of a bispecific nanobody conjugate broadly neutralizes diverse SARS-CoV-2 variants and structural basis for its broad neutralization.
Plos Pathog., 19, 2023
|
|
8H5T
 
 | |
5Z2C
 
 | | Crystal structure of ALPK-1 N-terminal domain in complex with ADP-heptose | | Descriptor: | Alpha-protein kinase 1, [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4S,5S,6R)-6-[(1S)-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl] hydrogen phosphate | | Authors: | Ding, J, She, Y, Shao, F. | | Deposit date: | 2018-01-02 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Alpha-kinase 1 is a cytosolic innate immune receptor for bacterial ADP-heptose.
Nature, 561, 2018
|
|
8J6K
 
 | | Crystal structure of pro-interleukin-18 and caspase-4 complex | | Descriptor: | Arginine ADP-riboxanase OspC3, Caspase-4 subunit p10, Caspase-4 subunit p20, ... | | Authors: | Sun, Q, Hou, Y.J, Ding, J. | | Deposit date: | 2023-04-26 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Recognition and maturation of IL-18 by caspase-4 noncanonical inflammasome.
Nature, 624, 2023
|
|
7VCF
 
 | | Cryo-EM structure of Chlamydomonas TOC-TIC supercomplex | | Descriptor: | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wu, J, Yan, Z, Jin, Z, Zhang, Y. | | Deposit date: | 2021-09-02 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure of a TOC-TIC supercomplex spanning two chloroplast envelope membranes.
Cell, 185, 2022
|
|
7WGC
 
 | | Neutral Omicron Spike Trimer in complex with ACE2. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Cui, Z. | | Deposit date: | 2021-12-28 | | Release date: | 2022-06-22 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron.
Cell, 185, 2022
|
|
