2RNM
 
 | | Structure of The HET-s(218-289) prion in its amyloid form obtained by solid-state NMR | | Descriptor: | Small s protein | | Authors: | Wasmer, C, Lange, A, Van Melckebeke, H, Siemer, A, Riek, R, Meier, B.H. | | Deposit date: | 2008-01-24 | | Release date: | 2008-04-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Amyloid fibrils of the HET-s(218-289) prion form a beta solenoid with a triangular hydrophobic core
Science, 319, 2008
|
|
2UYM
 
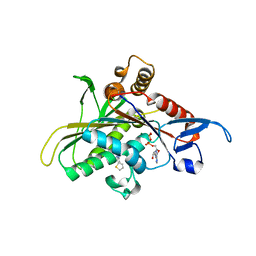 | |
2UYI
 
 | | Crystal structure of KSP in complex with ADP and thiophene containing inhibitor 33 | | Descriptor: | (5R)-N,N-DIETHYL-5-METHYL-2-[(THIOPHEN-2-YLCARBONYL)AMINO]-4,5,6,7-TETRAHYDRO-1-BENZOTHIOPHENE-3-CARBOXAMIDE, ADENOSINE-5'-DIPHOSPHATE, KINESIN-LIKE PROTEIN KIF11, ... | | Authors: | Lee, T.T. | | Deposit date: | 2007-04-07 | | Release date: | 2007-05-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synthesis and Sar of Thiophene Containing Kinesin Spindle Protein (Ksp) Inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
1FFQ
 
 | | CRYSTAL STRUCTURE OF CHITINASE A COMPLEXED WITH ALLOSAMIDIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, CHITINASE A | | Authors: | Papanikolau, Y, Tavlas, G, Vorgias, C.E, Petratos, K. | | Deposit date: | 2000-07-26 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | De novo purification scheme and crystallization conditions yield high-resolution structures of chitinase A and its complex with the inhibitor allosamidin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1FLR
 
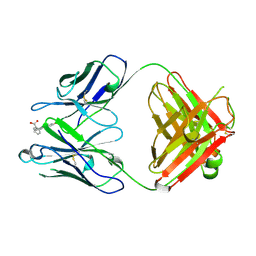 | | 4-4-20 FAB FRAGMENT | | Descriptor: | 2-(6-HYDROXY-3-OXO-3H-XANTHEN-9-YL)-BENZOIC ACID, 4-4-20 (IG*G2A=KAPPA=) FAB FRAGMENT | | Authors: | Whitlow, M. | | Deposit date: | 1995-01-19 | | Release date: | 1995-09-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 A structure of anti-fluorescein 4-4-20 Fab.
Protein Eng., 8, 1995
|
|
1EDQ
 
 | |
5KUX
 
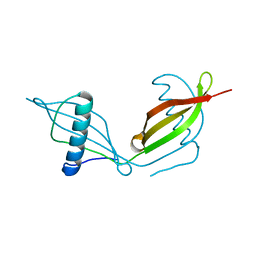 | |
5KUY
 
 | |
5MP6
 
 | | Structure of the Unliganded Fab from HIV-1 Neutralizing Antibody CAP248-2B that Binds to the gp120 C-terminus - gp41 Interface, at two Angstrom resolution. | | Descriptor: | CAP248-2B Heavy Chain, CAP248-2B Light Chain, SULFATE ION | | Authors: | Wibmer, C.K, Gorman, J, Kwong, P.D. | | Deposit date: | 2016-12-15 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.959 Å) | | Cite: | Structure and Recognition of a Novel HIV-1 gp120-gp41 Interface Antibody that Caused MPER Exposure through Viral Escape.
PLoS Pathog., 13, 2017
|
|
5M5Q
 
 | | COPS5(2-257) IN COMPLEX WITH A AZAINDOLE (COMPOUND 4) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-[(3~{R})-3-(1~{H}-benzimidazol-2-yl)morpholin-4-yl]-3-[2-(4-methyl-2-phenyl-phenyl)-1~{H}-pyrrolo[2,3-b]pyridin-3-yl]propan-1-one, COP9 signalosome complex subunit 5, ... | | Authors: | Renatus, M, Altmann, E. | | Deposit date: | 2016-10-22 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Azaindoles as Zinc-Binding Small-Molecule Inhibitors of the JAMM Protease CSN5.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5LXT
 
 | | Tubulin-Discodermolide complex | | Descriptor: | (+)-Discodermolide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Prota, A.E, Steinmetz, M.O. | | Deposit date: | 2016-09-22 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of Microtubule Stabilization by Discodermolide.
Chembiochem, 18, 2017
|
|
5LRQ
 
 | | BRD4 in complex with ERK5 inhibitor XMD8-92 | | Descriptor: | 2-{[2-ethoxy-4-(4-hydroxypiperidin-1-yl)phenyl]amino}-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain-containing protein 4 | | Authors: | Martin, M.P, Noble, M.E.M. | | Deposit date: | 2016-08-19 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of a novel orally bioavailable ERK5 inhibitor with selectivity over p38 alpha and BRD4.
Eur.J.Med.Chem., 178, 2019
|
|
5LXS
 
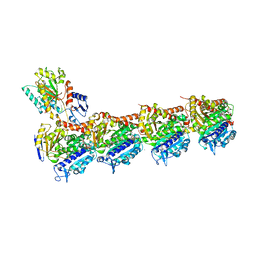 | | Tubulin-KS-1-199-32 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Prota, A.E, Steinmetz, M.O. | | Deposit date: | 2016-09-22 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Microtubule Stabilization by Discodermolide.
Chembiochem, 18, 2017
|
|
5M45
 
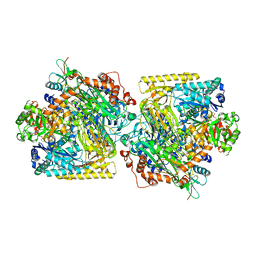 | | Structure of Acetone Carboxylase purified from Xanthobacter autotrophicus | | Descriptor: | 3,6,9,12,15-PENTAOXAHEPTADECAN-1-OL, ACETATE ION, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Kabasakal, B.V, Wells, J.N, Nwaobi, B.C, Eilers, B.J, Peters, J.W, Murray, J.W. | | Deposit date: | 2016-10-18 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural Basis for the Mechanism of ATP-Dependent Acetone Carboxylation.
Sci Rep, 7, 2017
|
|
5NNW
 
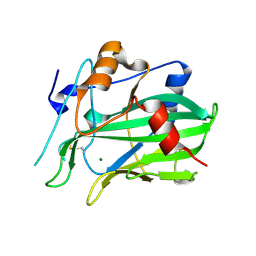 | | NLPPya in complex with glucosamine | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose, 25 kDa protein elicitor, MAGNESIUM ION | | Authors: | Podobnik, M, Anderluh, G, Lenarcic, T. | | Deposit date: | 2017-04-10 | | Release date: | 2017-12-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Eudicot plant-specific sphingolipids determine host selectivity of microbial NLP cytolysins.
Science, 358, 2017
|
|
5NO9
 
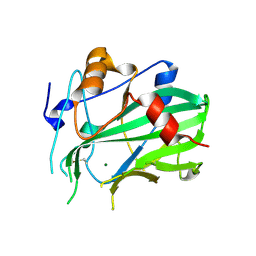 | | NLPPya in complex with mannosamine | | Descriptor: | 2-amino-2-deoxy-alpha-D-mannopyranose, 25 kDa protein elicitor, MAGNESIUM ION | | Authors: | Podobnik, M, Anderluh, G, Lenarcic, T. | | Deposit date: | 2017-04-11 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Eudicot plant-specific sphingolipids determine host selectivity of microbial NLP cytolysins.
Science, 358, 2017
|
|
7SQD
 
 | |
3DD5
 
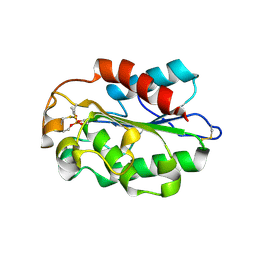 | | Glomerella cingulata E600-cutinase complex | | Descriptor: | Cutinase, DIETHYL PHOSPHONATE | | Authors: | Nyon, M.P, Rice, D.W, Berrisford, J.M, Hounslow, A.M, Moir, A.J.G, Huang, H, Nathan, S, Mahadi, N.M, Farah Diba, A.B, Craven, C.J. | | Deposit date: | 2008-06-05 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Catalysis by Glomerella cingulata Cutinase Requires Conformational Cycling between the Active and Inactive States of Its Catalytic Triad
J.Mol.Biol., 385, 2009
|
|
3DCN
 
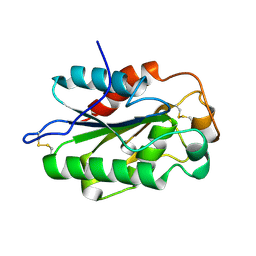 | | Glomerella cingulata apo cutinase | | Descriptor: | Cutinase | | Authors: | Nyon, M.P, Rice, D.W, Berrisford, J.M, Hounslow, A.M, Moir, A.J.G, Huang, H, Nathan, S, Mahadi, N.M, Farah Diba, A.B, Craven, C.J. | | Deposit date: | 2008-06-04 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalysis by Glomerella cingulata Cutinase Requires Conformational Cycling between the Active and Inactive States of Its Catalytic Triad
J.Mol.Biol., 385, 2009
|
|
3DEA
 
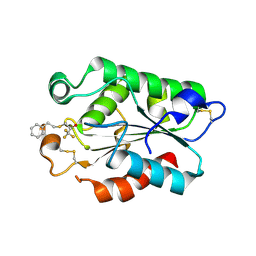 | | Glomerella cingulata PETFP-cutinase complex | | Descriptor: | 1,1,1-trifluoro-3-[(2-phenylethyl)sulfanyl]propan-2-one, Cutinase | | Authors: | Nyon, M.P, Rice, D.W, Berrisford, J.M, Hounslow, A.M, Moir, A.J.G, Huang, H, Nathan, S, Mahadi, N.M, Farah Diba, A.B, Craven, C.J. | | Deposit date: | 2008-06-09 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalysis by Glomerella cingulata Cutinase Requires Conformational Cycling between the Active and Inactive States of Its Catalytic Triad
J.Mol.Biol., 385, 2009
|
|
2CFX
 
 | | Structure of B.subtilis LrpC | | Descriptor: | HTH-TYPE TRANSCRIPTIONAL REGULATOR LRPC | | Authors: | Thaw, P, Rafferty, J.B. | | Deposit date: | 2006-02-24 | | Release date: | 2006-03-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insight Into Gene Transcriptional Regulation and Effector Binding by the Lrp/Asnc Family.
Nucleic Acids Res., 34, 2006
|
|
5OEW
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with glutamate and positive allosteric modulator BPAM538 | | Descriptor: | 4-cyclopropyl-7-(3-methoxyphenoxy)-2,3-dihydro-1$l^{6},2,4-benzothiadiazine 1,1-dioxide, ACETATE ION, GLUTAMIC ACID, ... | | Authors: | Larsen, A.P, Frydenvang, K.A, Kastrup, J.S. | | Deposit date: | 2017-07-10 | | Release date: | 2018-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 7-Phenoxy-Substituted 3,4-Dihydro-2H-1,2,4-benzothiadiazine 1,1-Dioxides as Positive Allosteric Modulators of alpha-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid (AMPA) Receptors with Nanomolar Potency.
J. Med. Chem., 61, 2018
|
|
2EWP
 
 | |
2DSN
 
 | | Crystal structure of T1 lipase | | Descriptor: | CALCIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Matsumura, H, Kai, Y. | | Deposit date: | 2006-07-03 | | Release date: | 2007-07-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel cation-pi interaction revealed by crystal structure of thermoalkalophilic lipase
Proteins, 70, 2007
|
|
2HAW
 
 | | Crystal structure of family II Inorganic pyrophosphatase in complex with PNP | | Descriptor: | CHLORIDE ION, FLUORIDE ION, GLYCEROL, ... | | Authors: | Fabrichniy, I.P, Lehtio, L, Oksanen, E, Goldman, A. | | Deposit date: | 2006-06-13 | | Release date: | 2006-11-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A trimetal site and substrate distortion in a family II inorganic pyrophosphatase
J.Biol.Chem., 282, 2007
|
|
