8GA7
 
 | |
4GU3
 
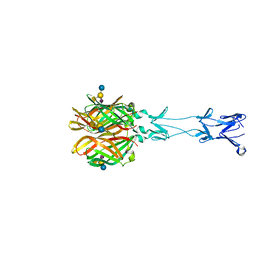 | |
2JWP
 
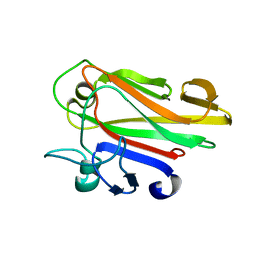 | | Malectin | | Descriptor: | Malectin | | Authors: | Schallus, T, Muhle-goll, C. | | Deposit date: | 2007-10-23 | | Release date: | 2008-08-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Malectin: A Novel Carbohydrate-binding Protein of the Endoplasmic Reticulum and a Candidate Player in the Early Steps of Protein N-Glycosylation
Mol.Cell.Biol., 19, 2008
|
|
4FQ2
 
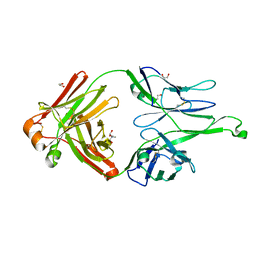 | | Crystal Structure of 10-1074 Fab | | Descriptor: | 1,2-ETHANEDIOL, Fab heavy chain, Fab light chain, ... | | Authors: | Scharf, L, Bjorkman, P.J. | | Deposit date: | 2012-06-24 | | Release date: | 2012-11-14 | | Last modified: | 2021-05-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2K46
 
 | | Xenopus laevis malectin complexed with nigerose (Glcalpha1-3Glc) | | Descriptor: | MGC80075 protein, alpha-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | Authors: | Schallus, T. | | Deposit date: | 2008-05-28 | | Release date: | 2008-08-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Malectin: A Novel Carbohydrate-binding Protein of the Endoplasmic Reticulum and a Candidate Player in the Early Steps of Protein N-Glycosylation
Mol.Cell.Biol., 19, 2008
|
|
4FQC
 
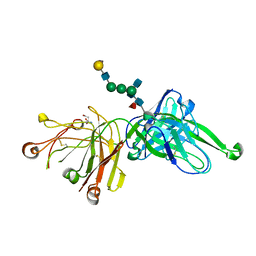 | | Crystal Structure of PGT121 Fab Bound to a complex-type sialylated N-glycan | | Descriptor: | Fab heavy chain, Fab light chain, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scharf, L, Bjorkman, P.J. | | Deposit date: | 2012-06-25 | | Release date: | 2012-11-14 | | Last modified: | 2021-05-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FQQ
 
 | |
7XP0
 
 | |
4EJN
 
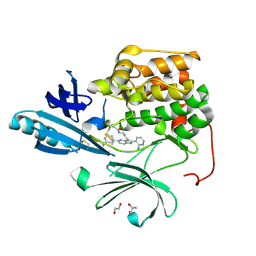 | | Crystal structure of autoinhibited form of AKT1 in complex with N-(4-(5-(3-acetamidophenyl)-2-(2-aminopyridin-3-yl)-3H-imidazo[4,5-b]pyridin-3-yl)benzyl)-3-fluorobenzamide | | Descriptor: | 1,2-ETHANEDIOL, 2-BUTANOL, N-(4-{5-[3-(acetylamino)phenyl]-2-(2-aminopyridin-3-yl)-3H-imidazo[4,5-b]pyridin-3-yl}benzyl)-3-fluorobenzamide, ... | | Authors: | Eathiraj, S. | | Deposit date: | 2012-04-06 | | Release date: | 2012-05-23 | | Last modified: | 2013-01-02 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Discovery and optimization of a series of 3-(3-phenyl-3H-imidazo[4,5-b]pyridin-2-yl)pyridin-2-amines: orally bioavailable, selective, and potent ATP-independent Akt inhibitors.
J.Med.Chem., 55, 2012
|
|
6K4L
 
 | |
4GU4
 
 | |
2LL4
 
 | |
7X0Y
 
 | |
7X0X
 
 | |
2LBO
 
 | |
7X88
 
 | |
6K4R
 
 | |
6K4K
 
 | |
2LL3
 
 | |
4HJU
 
 | |
4HJT
 
 | |
2NP9
 
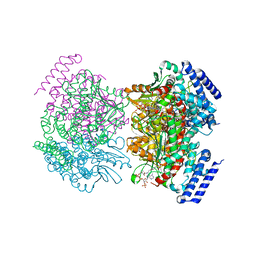 | | Crystal structure of a dioxygenase in the Crotonase superfamily | | Descriptor: | DpgC, OXYGEN MOLECULE, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL (3R)-4-({3-[(2-{[(3,5-DIHYDROXYPHENYL)ACETYL]AMINO}ETHYL)AMINO]-3-OXOPROPYL}AMINO)-3-HYDROXY-2,2-DIMETHYL-4-OXOBUTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Bruner, S.D, Widboom, P.F, Fielding, E.N. | | Deposit date: | 2006-10-26 | | Release date: | 2007-05-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for cofactor-independent dioxygenation in vancomycin biosynthesis.
Nature, 447, 2007
|
|
4HJS
 
 | |
4HJO
 
 | |
2M50
 
 | | Analysis of the structural and molecular basis of voltage-sensitive sodium channel inhibition by the spider toxin, Huwentoxin-IV (-TRTX-Hh2a). | | Descriptor: | Mu-theraphotoxin-Hh2a | | Authors: | Gibbs, A, Minassian, N, Flinspach, M, Wickenden, A. | | Deposit date: | 2013-02-12 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-29 | | Method: | SOLUTION NMR | | Cite: | Analysis of the Structural and Molecular Basis of Voltage-sensitive Sodium Channel Inhibition by the Spider Toxin Huwentoxin-IV ( mu-TRTX-Hh2a).
J.Biol.Chem., 288, 2013
|
|
