7TBH
 
 | | cryo-EM structure of MBP-KIX-apoferritin complex with peptide 7 | | Descriptor: | Isoform 2 of CREB-binding protein,Ferritin heavy chain, N-terminally processed, LEU-SER-ARG-ARG-PRO-SEP-TYR-ARG-LYS-ILE-LEU-ASN-ASP-LEU-SER-SER-ASP-ALA-PRO | | Authors: | Zhang, K, Horikoshi, N, Li, S, Powers, A, Hameedi, M, Pintilie, G, Chae, H, Khan, Y, Suomivuori, C, Dror, R, Sakamoto, K, Chiu, W, Wakatsuki, S. | | Deposit date: | 2021-12-22 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Cryo-EM, Protein Engineering, and Simulation Enable the Development of Peptide Therapeutics against Acute Myeloid Leukemia.
Acs Cent.Sci., 8, 2022
|
|
3MTL
 
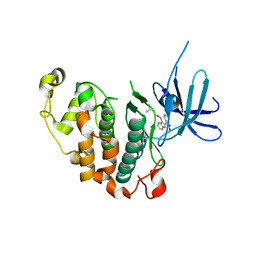 | | Crystal structure of the PCTAIRE1 kinase in complex with Indirubin E804 | | Descriptor: | (2Z,3E)-2,3'-BIINDOLE-2',3(1H,1'H)-DIONE 3-{O-[(3R)-3,4-DIHYDROXYBUTYL]OXIME}, Cell division protein kinase 16 | | Authors: | Krojer, T, Sharpe, T.D, Roos, A, Savitsky, P, Amos, A, Ayinampudi, V, Berridge, G, Fedorov, O, Keates, T, Phillips, C, Burgess-Brown, N, Zhang, Y, Pike, A.C.W, Muniz, J, Vollmar, M, Thangaratnarajah, C, Rellos, P, Ugochukwu, E, Filippakopoulos, P, Yue, W, Das, S, von Delft, F, Edwards, A, Arrowsmith, C.H, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and inhibitor specificity of the PCTAIRE-family kinase CDK16.
Biochem.J., 474, 2017
|
|
4QLB
 
 | | Structural Basis for the Recruitment of Glycogen Synthase by Glycogenin | | Descriptor: | GLYCEROL, Probable glycogen [starch] synthase, Protein GYG-1, ... | | Authors: | Zeqiraj, E, Judd, A, Sicheri, F. | | Deposit date: | 2014-06-11 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the recruitment of glycogen synthase by glycogenin.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2PMH
 
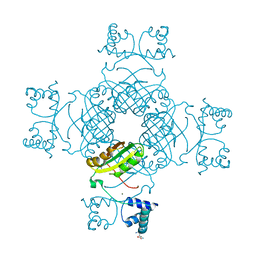 | | Crystal structure of Thr132Ala of ST1022 from Sulfolobus tokodaii | | Descriptor: | 150aa long hypothetical transcriptional regulator, GLUTAMINE, MAGNESIUM ION, ... | | Authors: | Kumarevel, T.S, Karthe, P, Nakano, N, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-22 | | Release date: | 2008-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of glutamine receptor protein from Sulfolobus tokodaii strain 7 in complex with its effector L-glutamine: implications of effector binding in molecular association and DNA binding
Nucleic Acids Res., 36, 2008
|
|
2PN6
 
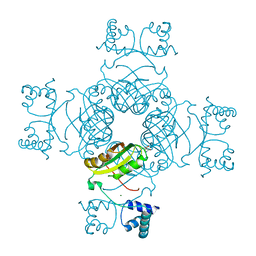 | | Crystal Structure of S32A of ST1022-Gln complex from Sulfolobus tokodaii | | Descriptor: | 150aa long hypothetical transcriptional regulator, GLUTAMINE, MAGNESIUM ION | | Authors: | Kumarevel, T.S, Karthe, P, Nakano, N, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-23 | | Release date: | 2008-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal structure of glutamine receptor protein from Sulfolobus tokodaii strain 7 in complex with its effector L-glutamine: implications of effector binding in molecular association and DNA binding
Nucleic Acids Res., 36, 2008
|
|
5X9P
 
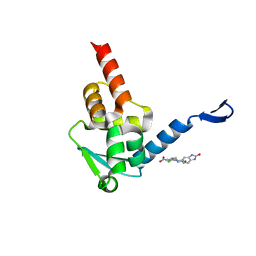 | | Crystal structure of the BCL6 BTB domain in complex with Compound 5 | | Descriptor: | 3-[[4-chloranyl-2-nitro-5-[(2-oxidanylidene-1,3-dihydrobenzimidazol-5-yl)amino]phenyl]amino]propanoic acid, B-cell lymphoma 6 protein | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-03-08 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Discovery of a novel B-cell lymphoma 6 (BCL6)-corepressor interaction inhibitor by utilizing structure-based drug design
Bioorg. Med. Chem., 25, 2017
|
|
5X9O
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 1a | | Descriptor: | 1,2-ETHANEDIOL, 5-[(2-chloranyl-4-nitro-phenyl)amino]-1,3-dihydrobenzimidazol-2-one, B-cell lymphoma 6 protein, ... | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-03-08 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Discovery of a novel B-cell lymphoma 6 (BCL6)-corepressor interaction inhibitor by utilizing structure-based drug design
Bioorg. Med. Chem., 25, 2017
|
|
5X4P
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 6 | | Descriptor: | 5-[(5-chloranylpyrimidin-4-yl)amino]-1,3-dihydroindol-2-one, B-cell lymphoma 6 protein | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach
J. Med. Chem., 60, 2017
|
|
5X4Q
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 7 | | Descriptor: | 1,2-ETHANEDIOL, 5-[[5-chloranyl-2-(pyridin-3-ylmethylamino)pyrimidin-4-yl]amino]-1,3-dihydroindol-2-one, B-cell lymphoma 6 protein, ... | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach
J. Med. Chem., 60, 2017
|
|
5X4M
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 1 | | Descriptor: | B-cell lymphoma 6 protein, N-phenyl-1,3,5-triazine-2,4-diamine | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach
J. Med. Chem., 60, 2017
|
|
5X4N
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 4 | | Descriptor: | 1,2-ETHANEDIOL, 5-chloro-N4-phenylpyrimidine-2,4-diamine, B-cell lymphoma 6 protein | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach
J. Med. Chem., 60, 2017
|
|
5X4O
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 5 | | Descriptor: | B-cell lymphoma 6 protein, N-methyl-N-{3-[({2-[(2-oxo-2,3-dihydro-1H-indol-5-yl)amino]-5-(trifluoromethyl)pyrimidin-4-yl}amino)methyl]pyridin-2-yl}methanesulfonamide | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach
J. Med. Chem., 60, 2017
|
|
8UWL
 
 | | 5-HT2AR bound to Lisuride in complex with a mini-Gq protein and an active-state stabilizing single-chain variable fragment (scFv16) obtained by cryo-electron microscopy (cryoEM) | | Descriptor: | 5-hydroxytryptamine receptor 2A, G protein subunit q (Gi2-mini-Gq chimera), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Barros-Alvarez, X, Kim, K, Panova, O, Roth, B.L, Skiniotis, G. | | Deposit date: | 2023-11-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | AlphaFold2 structures guide prospective ligand discovery.
Science, 384, 2024
|
|
5XCO
 
 | |
8V6U
 
 | | 5HT2AR-miniGq heterotrimer in complex with a novel agonist obtained from large scale docking | | Descriptor: | 4-{(3R)-1-[(1R)-1-(pyrimidin-2-yl)ethyl]piperidin-3-yl}phenol, 5-hydroxytryptamine receptor 2A, G protein alpha-subunit q (Gi2-mini-Gq chimera), ... | | Authors: | Gumpper, R.H, Wang, L, Kapolka, N, Skiniotis, G, Roth, B.L. | | Deposit date: | 2023-12-03 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | AlphaFold2 structures guide prospective ligand discovery.
Science, 384, 2024
|
|
2VKI
 
 | | Structure of the PDK1 PH domain K465E mutant | | Descriptor: | 3-PHOPSHOINOSITIDE DEPENDENT PROTEIN KINASE 1, GLYCEROL, SULFATE ION | | Authors: | Komander, D, Bayascas, J.R, Deak, M, Alessi, D.R, van Aalten, D.M.F. | | Deposit date: | 2007-12-19 | | Release date: | 2008-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mutation of the Pdk1 Ph Domain Inhibits Protein Kinase B/Akt, Leading to Small Size and Insulin Resistance.
Mol.Cell.Biol., 28, 2008
|
|
5X54
 
 | | Crystal structure of the Keap1 Kelch domain in complex with a tetrapeptide | | Descriptor: | ACE-GLU-TRP-TRP-TRP, ACETATE ION, Kelch-like ECH-associated protein 1 | | Authors: | Sogabe, S, Kadotani, A, Lane, W, Snell, G. | | Deposit date: | 2017-02-14 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a Kelch-like ECH-associated protein 1-inhibitory tetrapeptide and its structural characterization
Biochem. Biophys. Res. Commun., 486, 2017
|
|
5EZV
 
 | |
7ZBN
 
 | |
8IFJ
 
 | |
5C2T
 
 | | Crystal structure of Mitochondrial rhodoquinol-fumarate reductase from Ascaris suum with rhodoquinone-2 | | Descriptor: | 2-amino-5-[(2E)-3,7-dimethylocta-2,6-dien-1-yl]-3-methoxy-6-methylcyclohexa-2,5-diene-1,4-dione, Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | Deposit date: | 2015-06-16 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
Int J Mol Sci, 16, 2015
|
|
5G6V
 
 | | Crystal structure of the PCTAIRE1 kinase in complex with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-({[3-tert-butyl-1-(quinolin-6-yl)-1H-pyrazol-5-yl]carbamoyl}amino)-3-fluorophenoxy]-N-methylpyridine-2-carboxamide, CYCLIN-DEPENDENT KINASE 16 | | Authors: | Dixon-Clarke, S.E, Galan Bartual, S, Elkins, J, Savitsky, P, Kopec, J, Mackenzie, A, Tallant, C, Heroven, C, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2016-08-16 | | Release date: | 2016-11-23 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and inhibitor specificity of the PCTAIRE-family kinase CDK16.
Biochem.J., 474, 2017
|
|
5B04
 
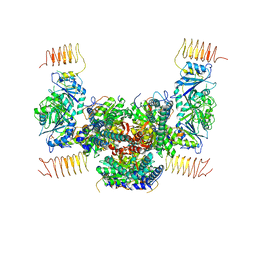 | | Crystal structure of the eukaryotic translation initiation factor 2B from Schizosaccharomyces pombe | | Descriptor: | PHOSPHATE ION, Probable translation initiation factor eIF-2B subunit beta, Probable translation initiation factor eIF-2B subunit delta, ... | | Authors: | Kashiwagi, K, Ito, T, Yokoyama, S. | | Deposit date: | 2015-10-27 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.994 Å) | | Cite: | Crystal structure of eukaryotic translation initiation factor 2B
Nature, 531, 2016
|
|
3VA2
 
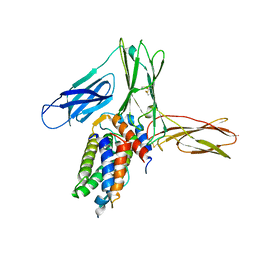 | | Crystal structure of human Interleukin-5 in complex with its alpha receptor | | Descriptor: | Interleukin-5, Interleukin-5 receptor subunit alpha | | Authors: | Kusano, S, Kukimoto-Niino, M, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2011-12-28 | | Release date: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Structural basis of interleukin-5 dimer recognition by its alpha receptor
Protein Sci., 21, 2012
|
|
6JP2
 
 | |
