3NVQ
 
 | | Molecular mechanism of guidance cue recognition | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-C1, ... | | Authors: | Juo, Z, Liu, H, Shim, A, Focia, P, Chen, X, Garcia, C, He, X. | | Deposit date: | 2010-07-08 | | Release date: | 2010-09-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Semaphorin-Plexin Recognition and Viral Mimicry from Sema7A and A39R Complexes with PlexinC1.
Cell(Cambridge,Mass.), 142, 2010
|
|
5XNP
 
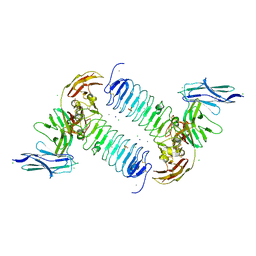 | | Crystal structures of human SALM5 in complex with human PTPdelta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Liu, H, Lin, Z, Xu, F. | | Deposit date: | 2017-05-24 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.729 Å) | | Cite: | Structural basis of SALM5-induced PTP delta dimerization for synaptic differentiation
Nat Commun, 9, 2018
|
|
5YJH
 
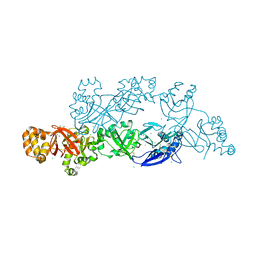 | | Structural insights into periostin functions | | Descriptor: | CALCIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Liu, H, Liu, J, Xu, F. | | Deposit date: | 2017-10-10 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.957 Å) | | Cite: | Structural characterizations of human periostin dimerization and cysteinylation.
FEBS Lett., 592, 2018
|
|
5YJG
 
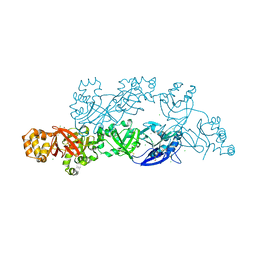 | | Structural insights into periostin functions | | Descriptor: | CALCIUM ION, CHLORIDE ION, CYSTEINE, ... | | Authors: | Liu, H, Liu, J, Xu, F. | | Deposit date: | 2017-10-10 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural characterizations of human periostin dimerization and cysteinylation.
FEBS Lett., 592, 2018
|
|
5XGP
 
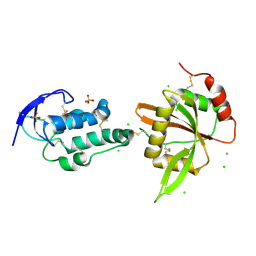 | | structure of Sizzled from Xenopus laevis at 2.08 angstroms resolution | | Descriptor: | CHLORIDE ION, SULFATE ION, Secreted Xwnt8 inhibitor sizzled | | Authors: | Liu, H, Li, Z, Xu, F. | | Deposit date: | 2017-04-15 | | Release date: | 2017-08-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.077 Å) | | Cite: | The crystal structure of full-length Sizzled from Xenopus laevis yields insights into Wnt-antagonistic function of secreted Frizzled-related proteins
J. Biol. Chem., 292, 2017
|
|
5Y4M
 
 | | Discoidin domain of human CASPR2 | | Descriptor: | 1,2-ETHANEDIOL, human CASPR2 Disc domain | | Authors: | Liu, H, Xu, F, Zhang, J, Liang, W. | | Deposit date: | 2017-08-04 | | Release date: | 2018-08-08 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural mapping of hot spots within human CASPR2 discoidin domain for autoantibody recognition.
J. Autoimmun., 96, 2019
|
|
4Q5Y
 
 | | Crystal structure of extended-Tudor 10-11 of Drosophila melanogaster | | Descriptor: | Maternal protein tudor | | Authors: | Liu, H, Ren, R, Wang, W, Wang, M, Yang, N, Dong, Y, Gong, W, Lehmann, R, Xu, R.M. | | Deposit date: | 2014-04-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and domain organization of Drosophila Tudor
Cell Res., 24, 2014
|
|
6L2L
 
 | |
6L2M
 
 | |
8I4L
 
 | | Capsid structure of the Cyanophage P-SCSP1u | | Descriptor: | The capsid protein(gp 19) of P-SCSP1u | | Authors: | Liu, H, Dang, S. | | Deposit date: | 2023-01-19 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Cryo-EM structure of cyanophage P-SCSP1u offers insights into DNA gating and evolution of T7-like viruses.
Nat Commun, 14, 2023
|
|
8I4M
 
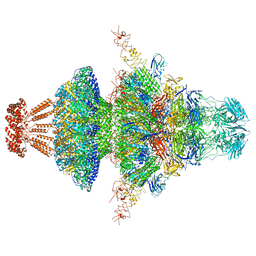 | | Portal-tail complex structure of the Cyanophage P-SCSP1u | | Descriptor: | Adaptor protein(gp22) of the cyanophage P-SCSP1u, Fiber protein(gp 28) of the cyanophage P-SCSP1u, Nozzle protein(gp 23) of the cyanophage P-SCSP1u, ... | | Authors: | Liu, H, Dang, S. | | Deposit date: | 2023-01-19 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Cryo-EM structure of cyanophage P-SCSP1u offers insights into DNA gating and evolution of T7-like viruses.
Nat Commun, 14, 2023
|
|
5ZVS
 
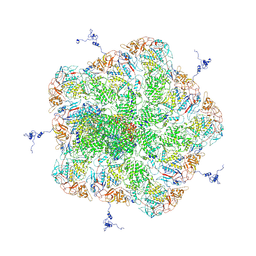 | |
5ZVT
 
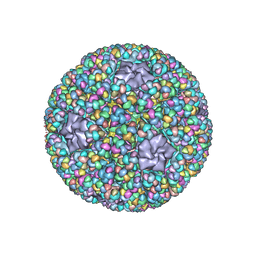 | | Structure of RNA polymerase complex and genome within a dsRNA virus provides insights into the mechanisms of transcription and assembly | | Descriptor: | C-terminus of outer capsid protein VP5, Core protein VP6, MYRISTIC ACID, ... | | Authors: | Liu, H, Fang, Q, Cheng, L. | | Deposit date: | 2018-05-12 | | Release date: | 2018-07-04 | | Last modified: | 2018-07-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of RNA polymerase complex and genome within a dsRNA virus provides insights into the mechanisms of transcription and assembly.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6AVS
 
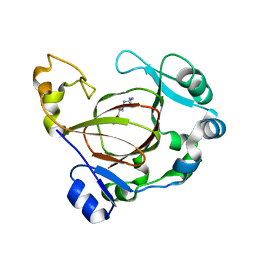 | | Complex structure of JMJD5 and Symmetric Monomethyl-Arginine (MMA) | | Descriptor: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, Lysine-specific demethylase 8, ZINC ION | | Authors: | Lee, S, Liu, H, Wang, Y, Dai, S, Zhang, G. | | Deposit date: | 2017-09-04 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Specific Recognition of Arginine Methylated Histone Tails by JMJD5 and JMJD7.
Sci Rep, 8, 2018
|
|
7U4L
 
 | | Crystal structure of human GPX4-U46C in complex with MAC-5576 | | Descriptor: | Phospholipid hydroperoxide glutathione peroxidase, thiophene-2-carbaldehyde | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Polychronidou, V, Soni, R.K, Xia, X, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4K
 
 | | Crystal structure of human GPX4-U46C-R152H in complex with ML162 | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-N-(3-chloro-4-methoxyphenyl)-N-[(1R)-2-oxo-2-[(2-phenylethyl)amino]-1-(thiophen-2-yl)ethyl]acetamide, Phospholipid hydroperoxide glutathione peroxidase | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Xia, X, Soni, R.K, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4N
 
 | | Crystal structure of human GPX4-U46C in complex with RSL3 | | Descriptor: | Phospholipid hydroperoxide glutathione peroxidase, methyl (1S,3R)-2-(chloroacetyl)-1-[4-(methoxycarbonyl)phenyl]-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indole-3-carboxylate | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Xia, X, Soni, R.K, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4M
 
 | | Crystal structure of human GPX4-U46C in complex with LOC1886 | | Descriptor: | 1,2-ETHANEDIOL, 4-methoxy-1H-indole-2-carbaldehyde, Phospholipid hydroperoxide glutathione peroxidase | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Polychronidou, V, Soni, R.K, Xia, X, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-28 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4J
 
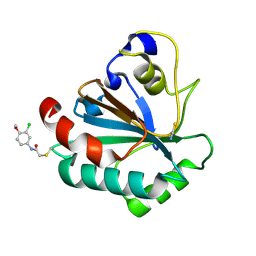 | | Crystal structure of human GPX4-U46C-R152H in complex with TMT10 | | Descriptor: | Phospholipid hydroperoxide glutathione peroxidase, THIOCYANATE ION, ~{N}-(3-chloranyl-4-methoxy-phenyl)ethanamide | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Polychronidou, V, Soni, R.K, Xia, X, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4I
 
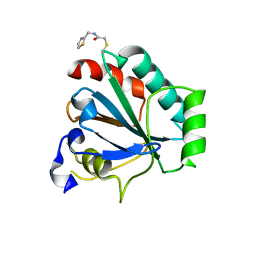 | | Crystal structure of human GPX4-U46C-R152H in complex with CDS9 | | Descriptor: | 2-bromo-N-[(thiophen-2-yl)methyl]acetamide, Phospholipid hydroperoxide glutathione peroxidase, THIOCYANATE ION | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Polychronidou, V, Soni, R.K, Xia, X, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
3ZFV
 
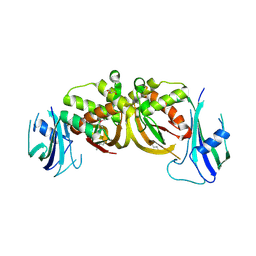 | | Crystal structure of an archaeal CRISPR-associated Cas6 nuclease | | Descriptor: | CRISPR-ASSOCIATED ENDORIBONUCLEASE CAS6 1, GLYCEROL | | Authors: | Reeks, J, Liu, H, White, M.F, Naismith, J.H. | | Deposit date: | 2012-12-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a Dimeric Crenarchaeal Cas6 Enzyme with an Atypical Active Site for Crispr RNA Processing
Biochem.J., 452, 2013
|
|
3B3Q
 
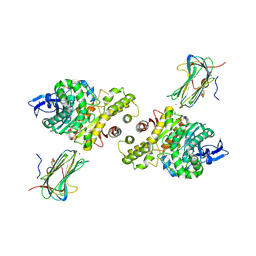 | | Crystal structure of a synaptic adhesion complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Chen, X, Liu, H, Shim, A, Focia, P, He, X. | | Deposit date: | 2007-10-22 | | Release date: | 2008-01-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for synaptic adhesion mediated by neuroligin-neurexin interactions.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2ALW
 
 | | Golgi alpha-mannosidase II complex with Noeuromycin | | Descriptor: | (2S,3S,4R,5R)-2,3,4-TRIHYDROXY-5-HYDROXYMETHYL-PIPERIDINE, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Bols, M.B, Liu, H, Rose, D.R. | | Deposit date: | 2005-08-08 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The role of the active site Zn in the catalytic mechanism of the GH38 Golgi alpha-mannosidase II: implications from noeuromycin inhibition
Biocatal.Biotransfor., 34, 2006
|
|
4UX5
 
 | | Structure of DNA complex of PCG2 | | Descriptor: | 5'-D(*CP*AP*AP*TP*GP*AP*CP*GP*CP*GP*TP*AP*AP*GP)-3', 5'-D(*CP*TP*TP*AP*CP*GP*CP*GP*TP*CP*AP*TP*TP*GP)-3', TRANSCRIPTION FACTOR MBP1 | | Authors: | Liu, J, Huang, J, Zhao, Y, Liu, H, Wang, D, Yang, J, Zhao, W, Taylor, I.A, Peng, Y. | | Deposit date: | 2014-08-19 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of DNA Recognition by Pcg2 Reveals a Novel DNA Binding Mode for Winged Helix-Turn-Helix Domains.
Nucleic Acids Res., 43, 2015
|
|
6XC4
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody CC12.3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.3 heavy chain, CC12.3 light chain, ... | | Authors: | Yuan, M, Liu, H, Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Structural basis of a shared antibody response to SARS-CoV-2.
Science, 369, 2020
|
|
