2IE3
 
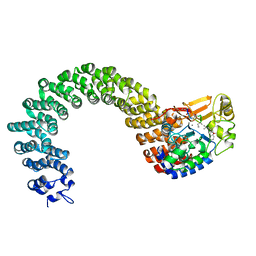 | | Structure of the Protein Phosphatase 2A Core Enzyme Bound to Tumor-inducing Toxins | | Descriptor: | MANGANESE (II) ION, Protein Phosphatase 2, regulatory subunit A (PR 65), ... | | Authors: | Xing, Y, Xu, Y, Chen, Y, Jeffrey, P.D, Chao, Y, Shi, Y. | | Deposit date: | 2006-09-17 | | Release date: | 2006-11-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Protein Phosphatase 2A Core Enzyme Bound to Tumor-Inducing Toxins
Cell(Cambridge,Mass.), 127, 2006
|
|
9M3O
 
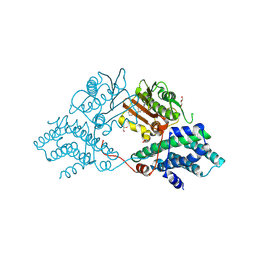 | |
9M3P
 
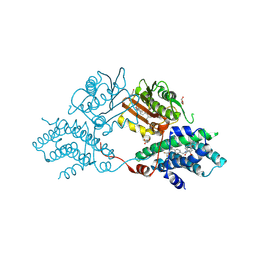 | |
9LSD
 
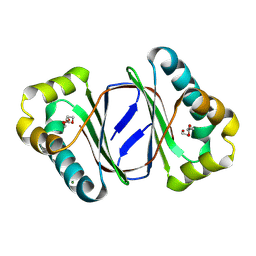 | |
6LPW
 
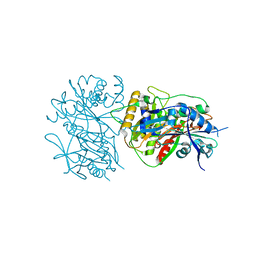 | |
7F6L
 
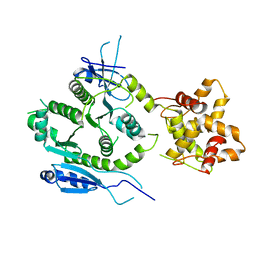 | | Crystal structure of human MUS81-EME2 complex | | Descriptor: | Crossover junction endonuclease MUS81, Probable crossover junction endonuclease EME2 | | Authors: | Hua, Z.K, Zhang, D.P, Yuan, C, Lin, Z.H. | | Deposit date: | 2021-06-25 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the human MUS81-EME2 complex.
Structure, 30, 2022
|
|
6LPV
 
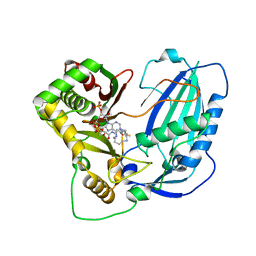 | |
4WXM
 
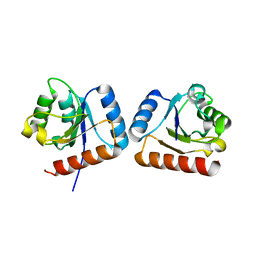 | | FleQ REC domain from Pseudomonas aeruginosa PAO1 | | Descriptor: | Transcriptional regulator FleQ | | Authors: | Su, T, Liu, S, Gu, L. | | Deposit date: | 2014-11-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The REC domain mediated dimerization is critical for FleQ from Pseudomonas aeruginosa to function as a c-di-GMP receptor and flagella gene regulator
J.Struct.Biol., 192, 2015
|
|
6O6W
 
 | | Solution structure of human myeloid-derived growth factor | | Descriptor: | Myeloid-derived growth factor | | Authors: | Bortnov, V, Tonelli, M, Lee, W, Markley, J.L, Mosher, D.F. | | Deposit date: | 2019-03-07 | | Release date: | 2019-11-13 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human myeloid-derived growth factor suggests a conserved function in the endoplasmic reticulum.
Nat Commun, 10, 2019
|
|
8ZCA
 
 | | Crystal structure of human CD47 ECD bound to Fab of Hu1C8 | | Descriptor: | 1C8 Fab heavy chain, 1C8 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, Z, Ding, J. | | Deposit date: | 2024-04-29 | | Release date: | 2025-05-07 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An anti-CD47 antibody binds to a distinct epitope in a novel metal ion-dependent manner to minimize cross-linking of red blood cells.
J.Biol.Chem., 2025
|
|
6VZA
 
 | | Crystal structure of cytochrome P450 NasF5053 Q65I-A86G mutant variant from Streptomyces sp. NRRL F-5053 in the cyclo-L-Trp-L-Pro-bound state | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Luo, Z, Jia, X, Sun, C, Qu, X, Kobe, B. | | Deposit date: | 2020-02-28 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Molecular basis of regio- and stereo-specificity in biosynthesis of bacterial heterodimeric diketopiperazines.
Nat Commun, 11, 2020
|
|
6VXV
 
 | | Crystal structure of cyclo-L-Trp-L-Pro-bound cytochrome P450 NasF5053 from Streptomyces sp. NRRL F-5053 | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Luo, Z, Jia, X, Sun, C, Qu, X, Kobe, B. | | Deposit date: | 2020-02-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of regio- and stereo-specificity in biosynthesis of bacterial heterodimeric diketopiperazines.
Nat Commun, 11, 2020
|
|
6W0S
 
 | | Crystal structure of substrate free cytochrome P450 NasF5053 from Streptomyces sp. NRRL F-5053 | | Descriptor: | BROMIDE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Luo, Z, Jia, X, Sun, C, Qu, X, Kobe, B. | | Deposit date: | 2020-03-02 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of regio- and stereo-specificity in biosynthesis of bacterial heterodimeric diketopiperazines.
Nat Commun, 11, 2020
|
|
6VZB
 
 | | Crystal structure of cytochrome P450 NasF5053 S284A-V288A mutant variant from Streptomyces sp. NRRL F-5053 in the cyclo-L-Trp-L-Pro-bound state | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Luo, Z, Jia, X, Sun, C, Qu, X, Kobe, B. | | Deposit date: | 2020-02-28 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Molecular basis of regio- and stereo-specificity in biosynthesis of bacterial heterodimeric diketopiperazines.
Nat Commun, 11, 2020
|
|
6TYY
 
 | | Hedgehog autoprocessing mutant D46H | | Descriptor: | Protein hedgehog | | Authors: | Li, H, Li, Z, Wang, C, Callahan, B.P. | | Deposit date: | 2019-08-09 | | Release date: | 2019-11-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | General Base Swap Preserves Activity and Expands Substrate Tolerance in Hedgehog Autoprocessing.
J.Am.Chem.Soc., 141, 2019
|
|
1B4W
 
 | |
9KHV
 
 | | Structure of DdmD dimer with ssDNA without nucleotide | | Descriptor: | DNA (5'-D(P*AP*AP*CP*AP*TP*TP*AP*CP*AP*AP*AP*A)-3'), DNA (5'-D(P*AP*CP*AP*TP*TP*AP*CP*AP*AP*AP*AP*T)-3'), Helicase/UvrB N-terminal domain-containing protein | | Authors: | Lin, Z.H, Gao, H.S, Liu, Y.S, Li, R.Y. | | Deposit date: | 2024-11-11 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | A gate-clamp mechanism for ssDNA translocation by DdmD in Vibrio cholerae plasmid defense.
Nucleic Acids Res., 53, 2025
|
|
9KI0
 
 | | structure of DdmD dimer with ssDNA with AGS | | Descriptor: | DNA (5'-D(P*CP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Helicase/UvrB N-terminal domain-containing protein, ... | | Authors: | Lin, Z.H, Gao, H.S, Liu, Y.S, Li, R.Y. | | Deposit date: | 2024-11-11 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | A gate-clamp mechanism for ssDNA translocation by DdmD in Vibrio cholerae plasmid defense.
Nucleic Acids Res., 53, 2025
|
|
9KHZ
 
 | | Structure of DdmD dimer with ssDNA with ADP bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*CP*CP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA (5'-D(P*CP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Lin, Z.H, Gao, H.S, Liu, Y.S, Li, R.Y. | | Deposit date: | 2024-11-11 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | A gate-clamp mechanism for ssDNA translocation by DdmD in Vibrio cholerae plasmid defense.
Nucleic Acids Res., 53, 2025
|
|
5JJU
 
 | | Crystal structure of Rv2837c complexed with 5'-pApA and 5'-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MANGANESE (II) ION, RNA (5'-R(P*AP*A)-3'), ... | | Authors: | Wang, F, He, Q, Liu, S, Gu, L. | | Deposit date: | 2016-04-25 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.312 Å) | | Cite: | Structural and biochemical insight into the mechanism of Rv2837c from Mycobacterium tuberculosis as a c-di-NMP phosphodiesterase
J.Biol.Chem., 291, 2016
|
|
8HO1
 
 | | Crystal structure of cytochrome P450 NasF5053 mutant F387G | | Descriptor: | CALCIUM ION, Cytochrome P450-F5053, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ma, B.D, Tian, W, Qu, X, Kong, X.D. | | Deposit date: | 2022-12-09 | | Release date: | 2023-04-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering the Substrate Specificity of a P450 Dimerase Enables the Collective Biosynthesis of Heterodimeric Tryptophan-Containing Diketopiperazines.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8HNZ
 
 | | Crystal structure of cytochrome P450 NasF5053 mutant E73S complexed with 6FCWP | | Descriptor: | (3~{S},8~{a}~{S})-3-[(5-fluoranyl-1~{H}-indol-3-yl)methyl]-2,3,6,7,8,8~{a}-hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, CALCIUM ION, Cytochrome P450-F5053, ... | | Authors: | Ma, B.D, Tian, W, Qu, X, Kong, X.D. | | Deposit date: | 2022-12-09 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineering the Substrate Specificity of a P450 Dimerase Enables the Collective Biosynthesis of Heterodimeric Tryptophan-Containing Diketopiperazines.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8HO0
 
 | | Crystal structure of cytochrome P450 NasF5053 mutant E73S complexed with 8FCWP | | Descriptor: | (3~{S},8~{a}~{S})-3-[(7-fluoranyl-1~{H}-indol-3-yl)methyl]-2,3,6,7,8,8~{a}-hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, CALCIUM ION, Cytochrome P450-F5053, ... | | Authors: | Ma, B.D, Tian, W, Qu, X, Kong, X.D. | | Deposit date: | 2022-12-09 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Engineering the Substrate Specificity of a P450 Dimerase Enables the Collective Biosynthesis of Heterodimeric Tryptophan-Containing Diketopiperazines.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8HNY
 
 | | Crystal structure of cytochrome P450 NasF5053 mutant E73S complexed with 5FCWP | | Descriptor: | (3~{S},8~{a}~{S})-3-[(4-fluoranyl-1~{H}-indol-3-yl)methyl]-2,3,6,7,8,8~{a}-hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, Cytochrome P450-F5053, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ma, B.D, Tian, W, Qu, X, Kong, X.D. | | Deposit date: | 2022-12-09 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineering the Substrate Specificity of a P450 Dimerase Enables the Collective Biosynthesis of Heterodimeric Tryptophan-Containing Diketopiperazines.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
9BCE
 
 | | Shewanella oneidensis LysR family regulator SO0839 regulatory domain | | Descriptor: | Transcriptional regulator LysR family | | Authors: | Han, S.R, Liang, H.H, Lin, Z.H, Fan, Y.L, Ye, K, Gao, H.G, Tao, Y.J, Jin, M. | | Deposit date: | 2024-04-08 | | Release date: | 2024-11-20 | | Last modified: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A bacterial transcription activator dedicated to the expression of the enzyme catalyzing the first committed step in fatty acid biosynthesis.
Nucleic Acids Res., 52, 2024
|
|
