6FYQ
 
 | |
7AFZ
 
 | | L1 metallo-b-lactamase with compound EBL-1306 | | Descriptor: | 3-[3-chloranyl-4-(methylsulfonylmethyl)phenyl]-7-propan-2-yl-1~{H}-indole-2-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J, Brem, J. | | Deposit date: | 2020-09-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Imitation of beta-lactam binding enables broad-spectrum metallo-beta-lactamase inhibitors.
Nat.Chem., 14, 2022
|
|
6YC8
 
 | | Crystal structure of KRED1-Pglu enzyme | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Di Pisa, F. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural insights into the desymmetrization of bulky 1,2-dicarbonyls through enzymatic monoreduction.
Bioorg.Chem., 108, 2021
|
|
3F2M
 
 | | Urate oxidase complexed with 8-azaxanthine at 150 MPa | | Descriptor: | 8-AZAXANTHINE, SODIUM ION, Uricase | | Authors: | Colloc'h, N, Girard, E, Kahn, R, Fourme, R. | | Deposit date: | 2008-10-30 | | Release date: | 2009-11-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function perturbation and dissociation of tetrameric urate oxidase by high hydrostatic pressure.
Biophys.J., 98, 2010
|
|
7QTK
 
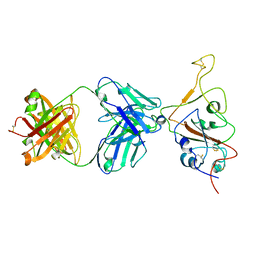 | | SARS-CoV-2 S Omicron Spike B.1.1.529 - RBD down - 1-P2G3 Fab (Local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, P2G3 Heavy Chain, P2G3 Light Chain, ... | | Authors: | Ni, D, Lau, K, Turelli, P, Fenwick, C, Perez, L, Pojer, F, Stahlberg, H, Pantaleo, G, Trono, D. | | Deposit date: | 2022-01-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Patient-derived monoclonal antibody neutralizes SARS-CoV-2 Omicron variants and confers full protection in monkeys.
Nat Microbiol, 7, 2022
|
|
7QTI
 
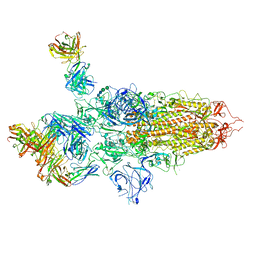 | | SARS-CoV-2 S Omicron Spike B.1.1.529 - 3-P2G3 and 1-P5C3 Fabs (Global) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, P2G3 Heavy Chain, ... | | Authors: | Ni, D, Lau, K, Turelli, P, Fenwick, C, Perez, L, Pojer, F, Stahlberg, H, Pantaleo, G, Trono, D. | | Deposit date: | 2022-01-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Patient-derived monoclonal antibody neutralizes SARS-CoV-2 Omicron variants and confers full protection in monkeys.
Nat Microbiol, 7, 2022
|
|
7QTJ
 
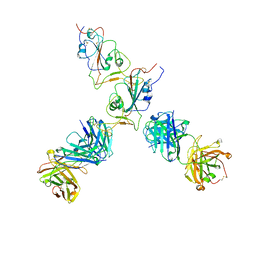 | | SARS-CoV-2 S Omicron Spike B.1.1.529 - RBD up - 1-P2G3 and 1-P5C3 Fabs (Local) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, P2G3 Heavy Chain, P2G3 Light Chain, ... | | Authors: | Ni, D, Lau, K, Turelli, P, Fenwick, C, Perez, L, Pojer, F, Stahlberg, H, Pantaleo, G, Trono, D. | | Deposit date: | 2022-01-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Patient-derived monoclonal antibody neutralizes SARS-CoV-2 Omicron variants and confers full protection in monkeys.
Nat Microbiol, 7, 2022
|
|
7AYJ
 
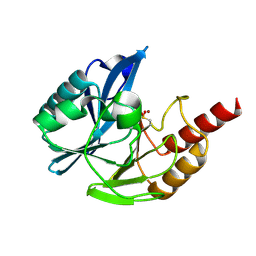 | | Metallo beta-lactamse Vim-1 with a sulfamoyl inhibitor. | | Descriptor: | 3-(4-fluorophenyl)-1-sulfamoyl-pyrrole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Farley, A.J.M, Ermolovich, I, Calvopina, K, Rabe, P, Brem, J, Schofield, C.J. | | Deposit date: | 2020-11-12 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural Basis of Metallo-beta-lactamase Inhibition by N -Sulfamoylpyrrole-2-carboxylates.
Acs Infect Dis., 7, 2021
|
|
5DOL
 
 | | Crystal structure of YabA amino-terminal domain from Bacillus subtilis | | Descriptor: | Initiation-control protein YabA | | Authors: | Cherrier, M.V, Bazin, A, Jameson, K.H, Wilkinson, A.J, Noirot-Gros, M.F, Terradot, L. | | Deposit date: | 2015-09-11 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Tetramerization and interdomain flexibility of the replication initiation controller YabA enables simultaneous binding to multiple partners.
Nucleic Acids Res., 44, 2016
|
|
6IRR
 
 | | Solution structure of DISC1/ATF4 complex | | Descriptor: | Disrupted in schizophrenia 1 homolog,Cyclic AMP-dependent transcription factor ATF-4 | | Authors: | Ye, F, Yu, C, Zhang, M. | | Deposit date: | 2018-11-14 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural interaction between DISC1 and ATF4 underlying transcriptional and synaptic dysregulation in an iPSC model of mental disorders.
Mol. Psychiatry, 2019
|
|
3GMD
 
 | | Structure-based design of 7-Azaindole-pyrrolidines as inhibitors of 11beta-Hydroxysteroid-Dehydrogenase type I | | Descriptor: | 2-methyl-3-{(3S)-1-[(1-pyridin-2-ylcyclopropyl)carbonyl]pyrrolidin-3-yl}-1H-pyrrolo[2,3-b]pyridine, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Valeur, E, Lepifre, F, Roche, D, Christmann-Franck, S, Hillertz, P, Musil, D. | | Deposit date: | 2009-03-13 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure-based design of 7-azaindole-pyrrolidine amides as inhibitors of 11 beta-hydroxysteroid dehydrogenase type I.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3DPA
 
 | |
3LC5
 
 | | Selective Benzothiophine Inhibitors of Factor IXa | | Descriptor: | 1-{4-[(R)-phenyl(3-phenyl-1,2,4-oxadiazol-5-yl)methoxy]-1-benzothiophen-2-yl}methanediamine, CALCIUM ION, Coagulation factor IX | | Authors: | Wang, S, Beck, R. | | Deposit date: | 2010-01-09 | | Release date: | 2010-02-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure Based Drug Design: Development of Potent and Selective Factor IXa (FIXa) Inhibitors.
J.Med.Chem., 53, 2010
|
|
6I0I
 
 | |
8ODW
 
 | | Crystal structure of LbmA Ox-ACP didomain in complex with NADP and ethyl glycinate from the lobatamide PKS (Gynuella sunshinyii) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Francois, R.M.M, Fraley, A.E, Piel, J, Weissman, K.J, Gruez, A. | | Deposit date: | 2023-03-09 | | Release date: | 2023-05-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Modular Oxime Formation by a trans-AT Polyketide Synthase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
5LI3
 
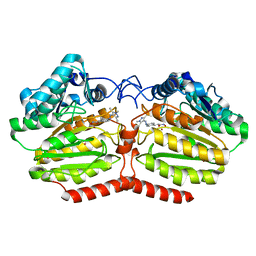 | | Crystal structure of HDAC-like protein from P. aeruginosa in complex with a photo-switchable inhibitor. | | Descriptor: | (2E)-N-hydroxy-3-{4-[(E)-(1,3,5-trimethyl-1H-pyrazol-4-yl)diazenyl]phenyl}prop-2-enamide, Acetoin utilization protein, POTASSIUM ION, ... | | Authors: | Kraemer, A, Meyer-Almes, F.J, Yildiz, O. | | Deposit date: | 2016-07-14 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Toward Photopharmacological Antimicrobial Chemotherapy Using Photoswitchable Amidohydrolase Inhibitors.
ACS Infect Dis, 3, 2017
|
|
9ETE
 
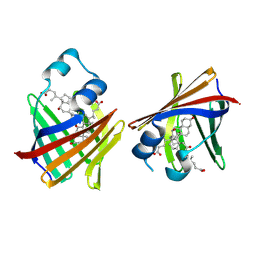 | | Crystal structure of recombinant chicken liver Bile Acid Binding Protein (cL-BABP) in complex with deoxycholic acid | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, Fatty acid-binding protein, liver | | Authors: | Tassone, G, Pozzi, C. | | Deposit date: | 2024-03-26 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Exploiting the bile acid binding protein as transporter of a Cholic Acid/Mirin bioconjugate for potential applications in liver cancer therapy.
Sci Rep, 14, 2024
|
|
9ETD
 
 | |
9ETF
 
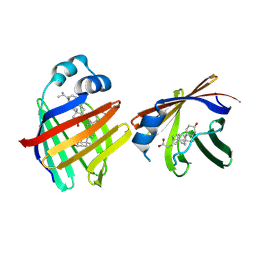 | | Crystal structure of recombinant chicken liver Bile Acid Binding Protein (cL-BABP) in complex with lithocholic acid | | Descriptor: | (3beta,5beta,14beta,17alpha)-3-hydroxycholan-24-oic acid, Fatty acid-binding protein, liver | | Authors: | Tassone, G, Pozzi, C. | | Deposit date: | 2024-03-26 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploiting the bile acid binding protein as transporter of a Cholic Acid/Mirin bioconjugate for potential applications in liver cancer therapy.
Sci Rep, 14, 2024
|
|
9ETC
 
 | |
9ETG
 
 | | Crystal structure of recombinant chicken liver Bile Acid Binding Protein (cL-BABP) in complex with CA-M11 | | Descriptor: | Fatty acid-binding protein, liver, [4-[(2-azanyl-4-oxidanylidene-1,3-thiazol-5-yl)methyl]phenyl] (4~{R})-4-[(3~{R},5~{R},7~{R},8~{R},9~{S},10~{S},12~{S},13~{R},14~{S},17~{R})-10,13-dimethyl-3,7,12-tris(oxidanyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1~{H}-cyclopenta[a]phenanthren-17-yl]pentanoate | | Authors: | Tassone, G, Pozzi, C, Maramai, S. | | Deposit date: | 2024-03-26 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploiting the bile acid binding protein as transporter of a Cholic Acid/Mirin bioconjugate for potential applications in liver cancer therapy.
Sci Rep, 14, 2024
|
|
6RKO
 
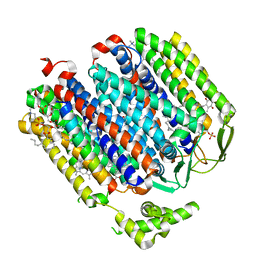 | | Cryo-EM structure of the E. coli cytochrome bd-I oxidase at 2.68 A resolution | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Cytochrome bd-I ubiquinol oxidase subunit 1, ... | | Authors: | Safarian, S, Hahn, A, Kuehlbrandt, W, Michel, H. | | Deposit date: | 2019-04-30 | | Release date: | 2019-10-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Active site rearrangement and structural divergence in prokaryotic respiratory oxidases.
Science, 366, 2019
|
|
6V6O
 
 | | EGFR(T790M/V948R) in complex with LN2380 | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, N-[3-({4-[4-(4-fluorophenyl)-2-(3-hydroxypropyl)-1H-imidazol-5-yl]pyridin-2-yl}amino)-4-methoxyphenyl]propanamide | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for EGFR Mutant Inhibition by Trisubstituted Imidazole Inhibitors.
J.Med.Chem., 63, 2020
|
|
6VHP
 
 | | Wild type EGFR in complex with LN2899 | | Descriptor: | Epidermal growth factor receptor, N-{3-[(4-{4-(4-fluorophenyl)-2-[(2-methoxyethyl)sulfanyl]-1H-imidazol-5-yl}pyridin-2-yl)amino]-4-methoxyphenyl}propanamide | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2020-01-10 | | Release date: | 2020-04-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural Basis for EGFR Mutant Inhibition by Trisubstituted Imidazole Inhibitors.
J.Med.Chem., 63, 2020
|
|
1A7G
 
 | |
