2I1R
 
 | | Novel Thiazolones as HCV NS5B Polymerase Inhibitors: Further Designs, Synthesis, SAR and X-ray Complex Structure | | Descriptor: | (5Z)-5-[(5-ETHYL-2-FURYL)METHYLENE]-2-{[(S)-(4-FLUOROPHENYL)(1H-TETRAZOL-5-YL)METHYL]AMINO}-1,3-THIAZOL-4(5H)-ONE, RNA-directed RNA polymerase (NS5B) (P68) | | Authors: | Yao, N, Yan, S. | | Deposit date: | 2006-08-14 | | Release date: | 2006-10-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel thiazolones as HCV NS5B polymerase allosteric inhibitors: Further designs, SAR, and X-ray complex structure.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2MP0
 
 | | Protein Phosphorylation upon a Fleeting Encounter | | Descriptor: | Glucose-specific phosphotransferase enzyme IIA component, PHOSPHITE ION, Phosphoenolpyruvate-protein phosphotransferase | | Authors: | Xing, Q, Yang, J, Huang, P, Zhang, W, Tang, C. | | Deposit date: | 2014-05-08 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Visualizing an ultra-weak protein-protein interaction in phosphorylation signaling.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3I19
 
 | |
4H47
 
 | | 1.9 angstrom CyPet structure at pH5.2 | | Descriptor: | ACETATE ION, Green fluorescent protein, SULFATE ION | | Authors: | Hu, X.-J, Liu, R. | | Deposit date: | 2012-09-17 | | Release date: | 2013-09-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights of the fluorescent states of CyPet
To be Published
|
|
7WNQ
 
 | | Cryo-EM structure of AtSLAC1 S59A mutant | | Descriptor: | Guard cell S-type anion channel SLAC1 | | Authors: | Sun, L, Liu, X, Li, Y. | | Deposit date: | 2022-01-19 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of the Arabidopsis guard cell anion channel SLAC1 suggests activation mechanism by phosphorylation.
Nat Commun, 13, 2022
|
|
4JNC
 
 | | Soluble Epoxide Hydrolase complexed with a carboxamide inhibitor | | Descriptor: | 1-[4-methyl-6-(methylamino)-1,3,5-triazin-2-yl]-N-[2-(trifluoromethyl)benzyl]piperidine-4-carboxamide, Bifunctional epoxide hydrolase 2 | | Authors: | Shewchuk, L.M. | | Deposit date: | 2013-03-15 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery of 1-(1,3,5-triazin-2-yl)piperidine-4-carboxamides as inhibitors of soluble epoxide hydrolase.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3GEX
 
 | |
1P5T
 
 | | Crystal Structure of Dok1 PTB Domain | | Descriptor: | Docking protein 1 | | Authors: | Shi, N, Ye, S, Liu, Y, Zhou, W, Ding, Y, Lou, Z, Qiang, B, Yuan, J, Rao, Z. | | Deposit date: | 2003-04-28 | | Release date: | 2004-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for the Specific Recognition of RET by the Dok1 Phosphotyrosine Binding Domain
J.BIOL.CHEM., 279, 2004
|
|
3HA8
 
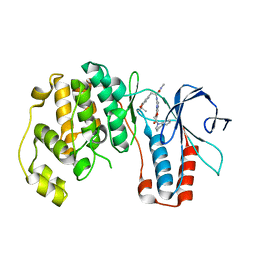 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/Compound 14b | | Descriptor: | Mitogen-activated protein kinase 14, N~2~-{4-[6-(3,4-dihydroquinolin-1(2H)-ylcarbonyl)-1H-benzimidazol-1-yl]-6-ethoxy-1,3,5-triazin-2-yl}-3-(2,2-dimethyl-4H-1,3-benzodioxin-6-yl)-N-methyl-L-alaninamide | | Authors: | Zhao, B, Clark, M.A. | | Deposit date: | 2009-05-01 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Design, synthesis and selection of DNA-encoded small-molecule libraries.
Nat.Chem.Biol., 5, 2009
|
|
3HA6
 
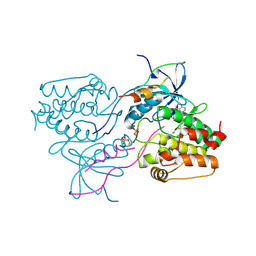 | | Crystal structure of aurora A in complex with TPX2 and compound 10 | | Descriptor: | N~2~-(3,4-dimethoxyphenyl)-N~4~-[2-(2-fluorophenyl)ethyl]-N~6~-quinolin-6-yl-1,3,5-triazine-2,4,6-triamine, Serine/threonine-protein kinase 6, Targeting protein for Xklp2 | | Authors: | Zhao, B, Clark, M.A. | | Deposit date: | 2009-05-01 | | Release date: | 2009-08-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Design, synthesis and selection of DNA-encoded small-molecule libraries.
Nat.Chem.Biol., 5, 2009
|
|
7DDX
 
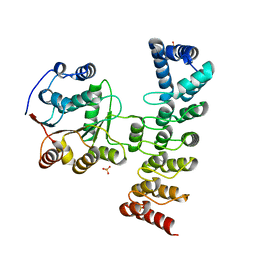 | | Crystal structure of KANK1 S1179F mutant in complex wtih eIF4A1 | | Descriptor: | Eukaryotic initiation factor 4A-I, GLYCEROL, KN motif and ankyrin repeat domains 1, ... | | Authors: | Pan, W, Xu, Y, Wei, Z. | | Deposit date: | 2020-10-30 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nephrotic-syndrome-associated mutation of KANK2 induces pathologic binding competition with physiological interactor KIF21A.
J.Biol.Chem., 297, 2021
|
|
2D2D
 
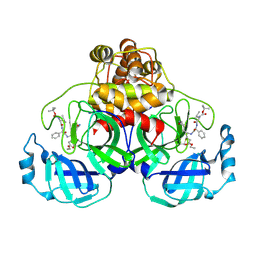 | | Crystal Structure Of SARS-CoV Mpro in Complex with an Inhibitor I2 | | Descriptor: | 3C-like proteinase, ETHYL (2E,4S)-4-[((2R)-2-{[N-(TERT-BUTOXYCARBONYL)-L-VALYL]AMINO}-2-PHENYLETHANOYL)AMINO]-5-[(3S)-2-OXOPYRROLIDIN-3-YL]PENT-2-ENOATE | | Authors: | Yang, H, Bartlam, M, Xue, X, Yang, K, Liang, W, Ding, Y, Rao, Z. | | Deposit date: | 2005-09-08 | | Release date: | 2005-09-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design of Wide-Spectrum Inhibitors Targeting Coronavirus Main Proteases.
Plos Biol., 3, 2005
|
|
7WZ5
 
 | | Larimichthys crocea IFNi | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interferon C | | Authors: | Chen, J.J. | | Deposit date: | 2022-02-17 | | Release date: | 2023-02-08 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural and Functional Characterization of a Fish Type I Subgroup d IFN Reveals Its Binding to Receptors.
J Immunol., 212, 2024
|
|
7BZJ
 
 | | The Discovery of Benzhydrol-Oxaborole Hybrid Derivatives as Leucyl-tRNA Synthetase Inhibitors | | Descriptor: | Leucine--tRNA ligase, [(1~{R},5~{R},6~{S},8~{R})-8-(6-aminopurin-9-yl)-4'-[(~{R})-oxidanyl-[4-(2-oxidanylidenepropylsulfanyl)phenyl]methyl]spiro[2,4,7-trioxa-3-boranuidabicyclo[3.3.0]octane-3,7'-7-boranuidabicyclo[4.3.0]nona-1(6),2,4-triene]-6-yl]methoxy-tris(oxidanyl)phosphanium | | Authors: | Liu, R.J, Li, H, Wang, E.D, Zhou, H. | | Deposit date: | 2020-04-28 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of benzhydrol-oxaborole derivatives as Streptococcus pneumoniae leucyl-tRNA synthetase inhibitors.
Bioorg.Med.Chem., 29, 2021
|
|
7DME
 
 | | Solution structure of human Aha1 | | Descriptor: | Activator of 90 kDa heat shock protein ATPase homolog 1 | | Authors: | Hu, H, Zhou, C, Zhang, N. | | Deposit date: | 2020-12-03 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Aha1 Exhibits Distinctive Dynamics Behavior and Chaperone-Like Activity.
Molecules, 26, 2021
|
|
7DMD
 
 | |
2MKV
 
 | |
2N2H
 
 | | Solution structure of Sds3 in complex with Sin3A | | Descriptor: | Paired amphipathic helix protein Sin3a, Sin3 histone deacetylase corepressor complex component SDS3 | | Authors: | Clark, M, Radhakrishnan, I. | | Deposit date: | 2015-05-08 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the assembly of the histone deacetylase-associated Sin3L/Rpd3L corepressor complex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2MUU
 
 | |
2Q6D
 
 | | Crystal structure of infectious bronchitis virus (IBV) main protease | | Descriptor: | Infectious bronchitis virus (IBV) main protease | | Authors: | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | Deposit date: | 2007-06-04 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
2Q6G
 
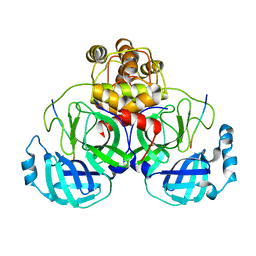 | | Crystal structure of SARS-CoV main protease H41A mutant in complex with an N-terminal substrate | | Descriptor: | Polypeptide chain, severe acute respiratory syndrome coronavirus (SARS-CoV) | | Authors: | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | Deposit date: | 2007-06-05 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
2REU
 
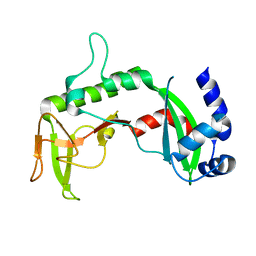 | | Crystal Structure of the C-terminal of Sau3AI fragment | | Descriptor: | MAGNESIUM ION, Type II restriction enzyme Sau3AI | | Authors: | Hu, X, Yu, F, Xu, C, He, J. | | Deposit date: | 2007-09-27 | | Release date: | 2008-09-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and function of C-terminal Sau3AI domain
Biochim.Biophys.Acta, 1794, 2009
|
|
2Q6F
 
 | | Crystal structure of infectious bronchitis virus (IBV) main protease in complex with a Michael acceptor inhibitor N3 | | Descriptor: | Infectious bronchitis virus (IBV) main protease, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | Deposit date: | 2007-06-05 | | Release date: | 2008-02-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
