7Y3W
 
 | |
8J7E
 
 | | Crystal structure of BRIL in complex with 1b3 Fab | | Descriptor: | Antibody 1b3 Fab Heavy chain, Antibody 1b3 Fab Light chain, Soluble cytochrome b562 | | Authors: | Zhong, Y.X, Guo, Q, Tao, Y.Y. | | Deposit date: | 2023-04-27 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A method for structure determination of GPCRs in various states.
Nat.Chem.Biol., 20, 2024
|
|
7WCY
 
 | | Crystal Structure of H-2Kb with Cryptosporidium parvum gp40/15 epitope | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-B alpha chain, ... | | Authors: | Wang, Y.L, Gao, M.H, Zhang, L.X, Fan, S.H. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural Analyses of a Dominant Cryptosporidium parvum Epitope Presented by H-2K b Offer New Options To Combat Cryptosporidiosis.
Mbio, 14, 2023
|
|
7YSU
 
 | | Cryo-EM Structure of FGF23-FGFR3c-aKlotho-HS Quaternary Complex | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, COPPER (II) ION, Fibroblast growth factor 23, ... | | Authors: | Mohammadi, M, Chen, L. | | Deposit date: | 2022-08-13 | | Release date: | 2023-04-19 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for FGF hormone signalling.
Nature, 618, 2023
|
|
7YSH
 
 | | Cryo-EM Structure of FGF23-FGFR1c-aKlotho-HS Quaternary Complex | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, COPPER (II) ION, Fibroblast growth factor 23, ... | | Authors: | Mohammadi, M, Chen, L. | | Deposit date: | 2022-08-12 | | Release date: | 2023-04-19 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural basis for FGF hormone signalling.
Nature, 618, 2023
|
|
7YSW
 
 | | Cryo-EM Structure of FGF23-FGFR4-aKlotho-HS Quaternary Complex | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, COPPER (II) ION, Fibroblast growth factor 23, ... | | Authors: | Mohammadi, M, Chen, L. | | Deposit date: | 2022-08-13 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural basis for FGF hormone signalling.
Nature, 618, 2023
|
|
7VUE
 
 | | Structural insight of the molecular mechanism of cilofexor bound to FXR | | Descriptor: | 2-[3-[4-[[3-[2,6-bis(chloranyl)phenyl]-5-cyclopropyl-1,2-oxazol-4-yl]methoxy]-2-chloranyl-phenyl]-3-oxidanyl-azetidin-1-yl]pyridine-4-carboxylic acid, Bile acid receptor, Peptide from Nuclear receptor coactivator 2 | | Authors: | Jiang, L, Chen, Y.C. | | Deposit date: | 2021-11-02 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural insight into the molecular mechanism of cilofexor binding to the farnesoid X receptor.
Biochem.Biophys.Res.Commun., 595, 2022
|
|
5JLY
 
 | | Structure of Peroxiredoxin-1 from Schistosoma japonicum | | Descriptor: | Thioredoxin peroxidase-1 | | Authors: | Wu, Q, Huang, F, Zeng, D, Liu, X, Zhao, J, Wang, H, Peng, Y, Li, P, Li, Y. | | Deposit date: | 2016-04-27 | | Release date: | 2017-05-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.051 Å) | | Cite: | Crystal structure of Peroxiredoxin-1 from Schistosoma japonicum
To Be Published
|
|
7C4D
 
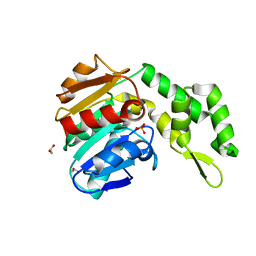 | | Marine microorganism esterase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Putative esterase, ... | | Authors: | Zhu, C.H, Wu, Y.K, Isupov, M.N. | | Deposit date: | 2020-05-16 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Insights into a Novel Esterase from the East Pacific Rise and Its Improved Thermostability by a Semirational Design.
J.Agric.Food Chem., 69, 2021
|
|
7DNJ
 
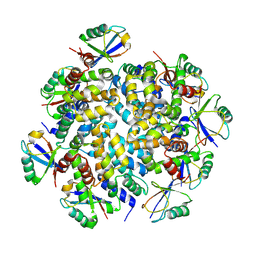 | | K63-polyUb MDA5CARDs complex | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, Ubiquitin | | Authors: | Song, B, Chen, Y, Luo, D.H, Zheng, J. | | Deposit date: | 2020-12-09 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ordered assembly of the cytosolic RNA-sensing MDA5-MAVS signaling complex via binding to unanchored K63-linked poly-ubiquitin chains.
Immunity, 54, 2021
|
|
7DNI
 
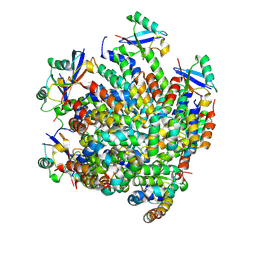 | | MDA5 CARDs-MAVS CARD polyUb complex | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, Mitochondrial antiviral-signaling protein, Ubiquitin | | Authors: | Song, B, Chen, Y, Luo, D.H, Zheng, J. | | Deposit date: | 2020-12-09 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Ordered assembly of the cytosolic RNA-sensing MDA5-MAVS signaling complex via binding to unanchored K63-linked poly-ubiquitin chains.
Immunity, 54, 2021
|
|
7DN9
 
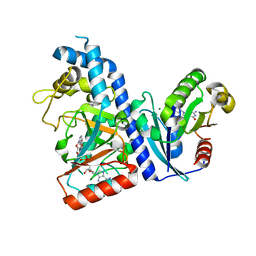 | |
7DN8
 
 | | Crystal structure of Salmonella effector SopF in complex with ARF1 | | Descriptor: | ADP-ribosylation factor 1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ding, J, Shao, F. | | Deposit date: | 2020-12-09 | | Release date: | 2021-12-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6084 Å) | | Cite: | ARF GTPases activate Salmonella effector SopF to ADP-ribosylate host V-ATPase and inhibit endomembrane damage-induced autophagy.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7DYR
 
 | |
7C4O
 
 | | Solution structure of the Orange domain from human protein HES1 | | Descriptor: | Transcription factor HES-1 | | Authors: | Fan, J.S, Nayak, A, Swaminathan, K. | | Deposit date: | 2020-05-18 | | Release date: | 2021-05-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Induction of Transcriptional Inhibitor Hairy and Enhancer of Split Homolog-1 and the Related Repression of Tumor-Suppressor Thioredoxin-Interacting Protein Are Important Components of Cell-Transformation Program Imposed by Oncogenic Kinase Nucleophosmin-Anaplastic Lymphoma Kinase.
Am J Pathol, 2022
|
|
7B92
 
 | |
7B9C
 
 | |
7B91
 
 | |
7F0S
 
 | |
3TG3
 
 | | Crystal structure of the MAPK binding domain of MKP7 | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity protein phosphatase 16 | | Authors: | Zhang, Y.Y, Liu, X, Wu, J.W, Wang, Z.X. | | Deposit date: | 2011-08-17 | | Release date: | 2012-03-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.675 Å) | | Cite: | A Distinct Interaction Mode Revealed by the Crystal Structure of the Kinase p38alpha with the MAPK Binding Domain of the Phosphatase MKP5.
Sci.Signal., 4, 2011
|
|
7W3U
 
 | | USP34 catalytic domain in complex with UbPA | | Descriptor: | Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 34, ZINC ION, ... | | Authors: | Xu, G.L, Ming, Z.H. | | Deposit date: | 2021-11-26 | | Release date: | 2022-06-01 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Structural Insights into the Catalytic Mechanism and Ubiquitin Recognition of USP34.
J.Mol.Biol., 434, 2022
|
|
7XJP
 
 | | Cryo-EM structure of EDS1 and SAG101 with ATP-APDR | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ADENOSINE-5-DIPHOSPHORIBOSE, ISOPROPYL ALCOHOL, ... | | Authors: | Huang, S.J, Jia, A.L, Han, Z.F, Chai, J.J. | | Deposit date: | 2022-04-18 | | Release date: | 2022-07-20 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | TIR-catalyzed ADP-ribosylation reactions produce signaling molecules for plant immunity.
Science, 377, 2022
|
|
7XME
 
 | | Structure of Influenza A virus polymerase basic protein 2 (PB2) with an azazindole derivative | | Descriptor: | (2~{S},3~{S})-3-[[5-dimethoxyphosphoryl-4-(5-fluoranyl-1~{H}-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-2-yl]amino]bicyclo[2.2.2]octane-2-carboxylic acid, IODIDE ION, Polymerase basic protein 2 | | Authors: | Zhang, Z. | | Deposit date: | 2022-04-25 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.521 Å) | | Cite: | Discovery of a novel azaindole derivatives targeting the influenza PB2 cap binding region
To Be Published
|
|
7W3R
 
 | | USP34 catalytic domain | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 34, ZINC ION | | Authors: | Xu, G.L, Ming, Z.H. | | Deposit date: | 2021-11-26 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Insights into the Catalytic Mechanism and Ubiquitin Recognition of USP34.
J.Mol.Biol., 434, 2022
|
|
7XOZ
 
 | | Crystal structure of RPPT-TIR | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase | | Authors: | Song, W, Jia, A, Huang, S, Chai, J. | | Deposit date: | 2022-05-02 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | TIR-catalyzed ADP-ribosylation reactions produce signaling molecules for plant immunity.
Science, 377, 2022
|
|
