7E9G
 
 | | Cryo-EM structure of Gi-bound metabotropic glutamate receptor mGlu2 | | Descriptor: | (1S,2S,5R,6S)-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylic acid, 1-butyl-3-chloranyl-4-(4-phenylpiperidin-1-yl)pyridin-2-one, DN13, ... | | Authors: | Lin, S, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2021-03-04 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of G i -bound metabotropic glutamate receptors mGlu2 and mGlu4.
Nature, 594, 2021
|
|
7E5Y
 
 | | Molecular basis for neutralizing antibody 2B11 targeting SARS-CoV-2 RBD | | Descriptor: | 2B11 Fab Heavy chain, 2B11 Fab Light chain, Spike protein S1 | | Authors: | Wu, H, Yu, F, Wang, Q.S, Zhou, H, Wang, W.W, Zhao, T, Pan, Y.B, Yang, X.M. | | Deposit date: | 2021-02-21 | | Release date: | 2021-06-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Screening of potent neutralizing antibodies against SARS-CoV-2 using convalescent patients-derived phage-display libraries.
Cell Discov, 7, 2021
|
|
4EGX
 
 | | Crystal structure of KIF1A CC1-FHA tandem | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Kinesin-like protein KIF1A | | Authors: | Yu, J, Huo, L, Yue, Y, Xu, T, Zhang, M, Feng, W. | | Deposit date: | 2012-04-02 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The CC1-FHA Tandem as a Central Hub for Controlling the Dimerization and Activation of Kinesin-3 KIF1A
Structure, 20, 2012
|
|
7DV8
 
 | | The crystal structure of rice immune receptor RGA5-HMA2. | | Descriptor: | Disease resistance protein RGA5 | | Authors: | Zhang, X, Liu, J.F. | | Deposit date: | 2021-01-12 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.447 Å) | | Cite: | A designer rice NLR immune receptor confers resistance to the rice blast fungus carrying noncorresponding avirulence effectors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3OQ1
 
 | | Crystal Structure of 11beta-Hydroxysteroid Dehydrogenase-1 (11b-HSD1) in Complex with Diarylsulfone Inhibitor | | Descriptor: | 3-(2-fluoroethyl)-4-({4-[(2S)-1,1,1-trifluoro-2-hydroxypropan-2-yl]phenyl}sulfonyl)benzonitrile, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wang, Z, Sudom, A, Walker, N.P. | | Deposit date: | 2010-09-02 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The synthesis and SAR of novel diarylsulfone 11beta-HSD1 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3QQ4
 
 | | Crystal structure of swine major histocompatibility complex class I SLA-1 0401 and identification of 2009 pandemic swine-origin influenza A H1N1 virus cytotoxic T lymphocyte epitope peptides | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, VP35 | | Authors: | Zhang, N, Qi, J, Gao, F, Pan, X, Chen, R, Li, Q, Chen, Z, Li, X, Xia, C, Gao, G.F. | | Deposit date: | 2011-02-15 | | Release date: | 2011-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Crystal structure of swine major histocompatibility complex class I SLA-1 0401 and identification of 2009 pandemic swine-origin influenza A H1N1 virus cytotoxic T lymphocyte epitope peptides.
J.Virol., 85, 2011
|
|
8Y6U
 
 | |
5EYC
 
 | | Crystal structure of c-Met in complex with naphthyridinone inhibitor 5 | | Descriptor: | 6-[(1~{R})-1-[8-fluoranyl-6-(3-methyl-1,2-oxazol-5-yl)-[1,2,4]triazolo[4,3-a]pyridin-3-yl]ethyl]-1,6-naphthyridin-5-one, Hepatocyte growth factor receptor | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of (R)-6-(1-(8-Fluoro-6-(1-methyl-1H-pyrazol-4-yl)-[1,2,4]triazolo[4,3-a]pyridin-3-yl)ethyl)-3-(2-methoxyethoxy)-1,6-naphthyridin-5(6H)-one (AMG 337), a Potent and Selective Inhibitor of MET with High Unbound Target Coverage and Robust In Vivo Antitumor Activity.
J.Med.Chem., 59, 2016
|
|
2HC5
 
 | | Solution NMR Structure of Protein yvyC from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR482. | | Descriptor: | Hypothetical protein yvyC | | Authors: | Eletsky, A, Liu, G, Atreya, H.S, Sukumaran, D.K, Wang, D, Cunningham, K, Janjua, H, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-15 | | Release date: | 2006-08-15 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structure of protein YvyC from Bacillus subtilis reveals unexpected structural similarity between two PFAM families.
Proteins, 76, 2009
|
|
5EYD
 
 | | Crystal structure of c-Met in complex with AMG 337 | | Descriptor: | 6-[(1~{R})-1-[8-fluoranyl-6-(1-methylpyrazol-4-yl)-[1,2,4]triazolo[4,3-a]pyridin-3-yl]ethyl]-3-(2-methoxyethoxy)-1,6-naphthyridin-5-one, Hepatocyte growth factor receptor | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of (R)-6-(1-(8-Fluoro-6-(1-methyl-1H-pyrazol-4-yl)-[1,2,4]triazolo[4,3-a]pyridin-3-yl)ethyl)-3-(2-methoxyethoxy)-1,6-naphthyridin-5(6H)-one (AMG 337), a Potent and Selective Inhibitor of MET with High Unbound Target Coverage and Robust In Vivo Antitumor Activity.
J.Med.Chem., 59, 2016
|
|
1CLK
 
 | | CRYSTAL STRUCTURE OF STREPTOMYCES DIASTATICUS NO.7 STRAIN M1033 XYLOSE ISOMERASE AT 1.9 A RESOLUTION WITH PSEUDO-I222 SPACE GROUP | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, XYLOSE ISOMERASE | | Authors: | Niu, L, Teng, M, Zhu, X, Gong, W. | | Deposit date: | 1999-04-29 | | Release date: | 2000-05-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of xylose isomerase from Streptomyces diastaticus no. 7 strain M1033 at 1.85 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
6D1N
 
 | |
8EFN
 
 | | Structure of Sp-STING3 from Stylophora pistillata coral in complex with 3',3'-cGAMP | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, Stimulator of interferon genes protein | | Authors: | Li, Y, Slavik, K.M, Morehouse, B.R, Mears, K, Kranzusch, P.J. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8EFM
 
 | | Structure of coral STING receptor from Stylophora pistillata in complex with 2',3'-cGAMP | | Descriptor: | SULFATE ION, Stimulator of interferon genes protein, cGAMP | | Authors: | Li, Y, Slavik, K.M, Morehouse, B.R, Mears, K, Kranzusch, P.J. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8EBP
 
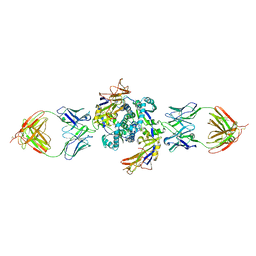 | | HMPV F dimer bound to RSV-199 Fab | | Descriptor: | Fusion glycoprotein F0, RSV-199 heavy chain protein, RSV-199 light chain protein | | Authors: | Wen, X, Jardetzky, T.S. | | Deposit date: | 2022-08-31 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Potent cross-neutralization of respiratory syncytial virus and human metapneumovirus through a structurally conserved antibody recognition mode.
Cell Host Microbe, 31, 2023
|
|
6D1P
 
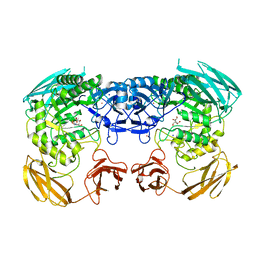 | | Apo structure of Bacteroides uniformis beta-glucuronidase 3 | | Descriptor: | GLYCEROL, Glycosyl hydrolases family 2, sugar binding domain protein, ... | | Authors: | Walton, W.G, Pellock, S.J, Redinbo, M.R. | | Deposit date: | 2018-04-12 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Three structurally and functionally distinct beta-glucuronidases from the human gut microbeBacteroides uniformis.
J. Biol. Chem., 293, 2018
|
|
7X08
 
 | | S protein of SARS-CoV-2 in complex with 2G1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-02-21 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Broad ultra-potent neutralization of SARS-CoV-2 variants by monoclonal antibodies specific to the tip of RBD.
Cell Discov, 8, 2022
|
|
6D41
 
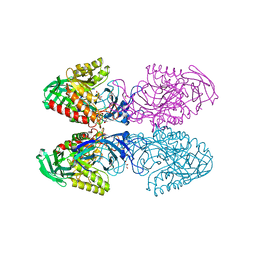 | | Bacteriodes uniformis beta-glucuronidase 1 bound to D-glucaro-1,5-lactone | | Descriptor: | (2S,3S,4S,5R)-3,4,5-trihydroxy-6-oxo-oxane-2-carboxylic acid, Beta-galactosidase/beta-glucuronidase, CHLORIDE ION, ... | | Authors: | Walton, W.G, Pellock, S.J, Redinbo, M.R. | | Deposit date: | 2018-04-17 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three structurally and functionally distinct beta-glucuronidases from the human gut microbeBacteroides uniformis.
J. Biol. Chem., 293, 2018
|
|
6D50
 
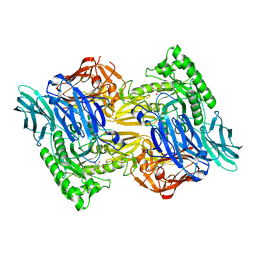 | | Bacteroides uniforms beta-glucuronidase 2 bound to D-glucaro-1,5-lactone | | Descriptor: | (2S,3S,4S,5R)-3,4,5-trihydroxy-6-oxo-oxane-2-carboxylic acid, CALCIUM ION, Glycosyl hydrolases family 2, ... | | Authors: | Walton, W.G, Pellock, S.J, Redinbo, M.R. | | Deposit date: | 2018-04-19 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three structurally and functionally distinct beta-glucuronidases from the human gut microbeBacteroides uniformis.
J. Biol. Chem., 293, 2018
|
|
6D89
 
 | |
5VZ2
 
 | |
5W18
 
 | | Staphylococcus aureus ClpP in complex with (S)-N-((2R,6S,8aS,14aS,20S,23aS)-2,6-dimethyl-5,8,14,19,23-pentaoxooctadecahydro-1H,5H,14H,19H-pyrido[2,1-i]dipyrrolo[2,1-c:2',1'-l][1]oxa[4,7,10,13]tetraazacyclohexadecin-20-yl)-3-phenyl-2-(3-phenylureido)propanamide | | Descriptor: | 9V7-PHE-SER-PRO-YCP-ALA-MP8, ATP-dependent Clp protease proteolytic subunit | | Authors: | Lee, R.E, Griffith, E.C. | | Deposit date: | 2017-06-02 | | Release date: | 2017-08-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Ureadepsipeptides as ClpP Activators.
Acs Infect Dis., 2019
|
|
8XAI
 
 | |
8GQ5
 
 | |
8GNJ
 
 | | Human SARM1 bounded with NMN and Nanobody-C6, Conformation 2 | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NAD(+) hydrolase SARM1, Nanobody-C6 | | Authors: | Cai, Y, Zhang, H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | A conformation-specific nanobody targeting the nicotinamide mononucleotide-activated state of SARM1.
Nat Commun, 13, 2022
|
|
