6PPG
 
 | |
5XBW
 
 | | The structure of BrlR | | Descriptor: | Probable transcriptional regulator | | Authors: | Wang, F, Qing, H, Gu, L. | | Deposit date: | 2017-03-21 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.109 Å) | | Cite: | BrlR from Pseudomonas aeruginosa is a receptor for both cyclic di-GMP and pyocyanin.
Nat Commun, 9, 2018
|
|
5XBI
 
 | |
5XBT
 
 | | The structure of BrlR bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Wang, F, Qing, H, Gu, L. | | Deposit date: | 2017-03-21 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | BrlR from Pseudomonas aeruginosa is a receptor for both cyclic di-GMP and pyocyanin.
Nat Commun, 9, 2018
|
|
3UCR
 
 | | Crystal structure of the immunoreceptor TIGIT IgV domain | | Descriptor: | CHLORIDE ION, T cell immunoreceptor with Ig and ITIM domains | | Authors: | Yin, J.P, Stengel, K.F, Rouge, L, Bazan, J.F, Wiesmann, C. | | Deposit date: | 2011-10-27 | | Release date: | 2012-03-14 | | Last modified: | 2012-04-25 | | Method: | X-RAY DIFFRACTION (2.627 Å) | | Cite: | Structure of TIGIT immunoreceptor bound to poliovirus receptor reveals a cell-cell adhesion and signaling mechanism that requires cis-trans receptor clustering.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UDW
 
 | | Crystal structure of the immunoreceptor TIGIT in complex with Poliovirus receptor (PVR/CD155/necl-5) D1 domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Poliovirus receptor, T cell immunoreceptor with Ig and ITIM domains | | Authors: | Rouge, L, Stengel, K.F, Yin, J.P, Bazan, F.J, Wiesmann, C. | | Deposit date: | 2011-10-28 | | Release date: | 2012-03-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Structure of TIGIT immunoreceptor bound to poliovirus receptor reveals a cell-cell adhesion and signaling mechanism that requires cis-trans receptor clustering.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2HDE
 
 | |
4MXW
 
 | | Structure of heterotrimeric lymphotoxin LTa1b2 bound to lymphotoxin beta receptor LTbR and anti-LTa Fab | | Descriptor: | Lymphotoxin-alpha, Lymphotoxin-beta, Tumor necrosis factor receptor superfamily member 3, ... | | Authors: | Sudhamsu, J, Yin, J.P, Hymowitz, S.G. | | Deposit date: | 2013-09-26 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Dimerization of LT beta R by LT alpha 1 beta 2 is necessary and sufficient for signal transduction.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4MXV
 
 | | Structure of Lymphotoxin alpha bound to anti-LTa Fab | | Descriptor: | Lymphotoxin-alpha, anti-Lymphotoxin alpha antibody heavy chain, anti-Lymphotoxin alpha antibody light chain | | Authors: | Yin, J.P, Hymowitz, S.G. | | Deposit date: | 2013-09-26 | | Release date: | 2013-11-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Dimerization of LT beta R by LT alpha 1 beta 2 is necessary and sufficient for signal transduction.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2QQO
 
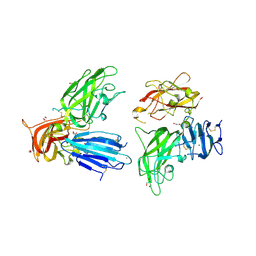 | | Crystal Structure of the a2b1b2 Domains from Human Neuropilin-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Appleton, B.A, Wiesmann, C. | | Deposit date: | 2007-07-26 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural studies of neuropilin/antibody complexes provide insights into semaphorin and VEGF binding
Embo J., 26, 2007
|
|
2QQI
 
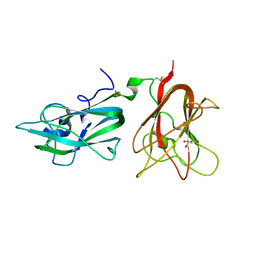 | |
2QQJ
 
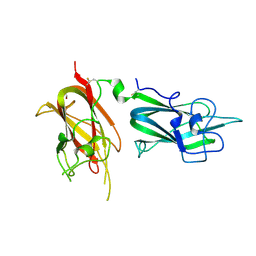 | |
2QQM
 
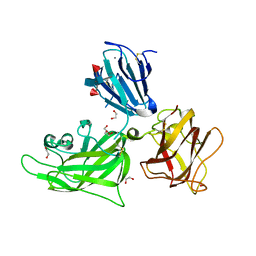 | | Crystal Structure of the a2b1b2 Domains from Human Neuropilin-1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Appleton, B.A, Wiesmann, C. | | Deposit date: | 2007-07-26 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural studies of neuropilin/antibody complexes provide insights into semaphorin and VEGF binding
Embo J., 26, 2007
|
|
2QQN
 
 | |
2QQK
 
 | |
2QQL
 
 | |
6U8X
 
 | | Crystal structure of DNMT3B-DNMT3L in complex with CpApG DNA | | Descriptor: | CpApG DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | Authors: | Gao, L, Song, J. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95063877 Å) | | Cite: | Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms.
Nat Commun, 11, 2020
|
|
6OQI
 
 | | CDK2 in complex with Cpd14 (5-fluoro-4-(4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl)-N-(5-(4-methylpiperazin-1-yl)pyridin-2-yl)pyrimidin-2-amine) | | Descriptor: | 5-fluoro-N-[5-(4-methylpiperazin-1-yl)pyridin-2-yl]-4-[(4S)-4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl]pyrimidin-2-amine, Cyclin-dependent kinase 2 | | Authors: | Murray, J.M. | | Deposit date: | 2019-04-26 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of a brain-penetrant CDK4/6 inhibitor for glioblastoma.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6OQO
 
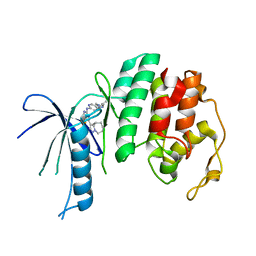 | | CDK6 in complex with Cpd24 N-(5-(6-ethyl-2,6-diazaspiro[3.3]heptan-2-yl)pyridin-2-yl)-5-fluoro-4-(4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl)pyrimidin-2-amine | | Descriptor: | Cyclin-dependent kinase 6, N-[5-(6-ethyl-2,6-diazaspiro[3.3]heptan-2-yl)pyridin-2-yl]-5-fluoro-4-[(4R)-4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl]pyrimidin-2-amine | | Authors: | Murray, J.M, Boenig, G.D.L. | | Deposit date: | 2019-04-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Design of a brain-penetrant CDK4/6 inhibitor for glioblastoma.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6U8P
 
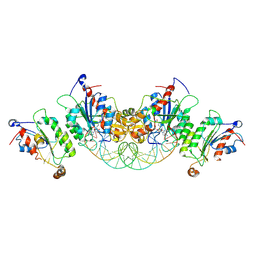 | | Crystal structure of DNMT3B-DNMT3L in complex with CpGpA DNA | | Descriptor: | CpGpA DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | Authors: | Gao, L, Song, J. | | Deposit date: | 2019-09-05 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms.
Nat Commun, 11, 2020
|
|
6U8V
 
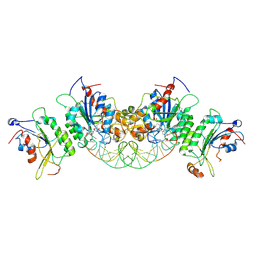 | | Crystal structure of DNMT3B-DNMT3L in complex with CpGpT DNA | | Descriptor: | CpGpT DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | Authors: | Gao, L, Zhang, Z.M, Song, J. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms.
Nat Commun, 11, 2020
|
|
6U8W
 
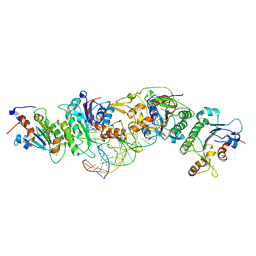 | | Crystal structure of DNMT3B(K777A)-DNMT3L in complex with CpGpT DNA | | Descriptor: | CpGpT DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | Authors: | Gao, L, Zhang, Z.M, Song, J. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.94891548 Å) | | Cite: | Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms.
Nat Commun, 11, 2020
|
|
6OQL
 
 | | CDK6 in complex with Cpd13 (R)-5-fluoro-4-(4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl)-N-(5-(4-methylpiperazin-1-yl)pyridin-2-yl)pyrimidin-2-amine | | Descriptor: | 5-fluoro-N-[5-(4-methylpiperazin-1-yl)pyridin-2-yl]-4-[(4R)-4-methyl-5,6,7,8-tetrahydro-4H-pyrazolo[1,5-a]azepin-3-yl]pyrimidin-2-amine, Cyclin-dependent kinase 6 | | Authors: | Murray, J.M. | | Deposit date: | 2019-04-26 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.707 Å) | | Cite: | Design of a brain-penetrant CDK4/6 inhibitor for glioblastoma.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6LON
 
 | | Crystal structure of HPSG | | Descriptor: | (2~{S})-2,3-bis(oxidanyl)propane-1-sulfonic acid, GLYCEROL, PFL2/glycerol dehydratase family glycyl radical enzyme, ... | | Authors: | Liu, J, Zhang, Y, Yuchi, Z. | | Deposit date: | 2020-01-06 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two radical-dependent mechanisms for anaerobic degradation of the globally abundant organosulfur compound dihydroxypropanesulfonate.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8TDR
 
 | |
