3Q3J
 
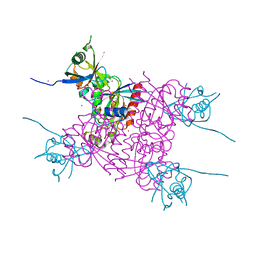 | | Crystal structure of plexin A2 RBD in complex with Rnd1 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Plexin-A2, ... | | Authors: | Wang, H, Tempel, W, Tong, Y, Guan, X, Shen, L, Buren, L, Zhang, N, Wernimont, A.K, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-21 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.971 Å) | | Cite: | Crystal structure of plexin A2 RBD in complex with Rnd1
to be published
|
|
2RIE
 
 | | Crystal structure of the trimeric neck and carbohydrate recognition domain of human surfactant protein D in complex with 2-deoxy-L-glycero-D-manno-heptose | | Descriptor: | 2-deoxy-beta-L-galacto-heptopyranose, CALCIUM ION, Pulmonary surfactant-associated protein D | | Authors: | Wang, H, Head, J, Kosma, P, Sheikh, S, McDonald, B, Smith, K, Cafarella, T, Seaton, B, Crouch, E. | | Deposit date: | 2007-10-10 | | Release date: | 2008-01-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of heptoses and the inner core of bacterial lipopolysaccharides by surfactant protein d.
Biochemistry, 47, 2008
|
|
3PFN
 
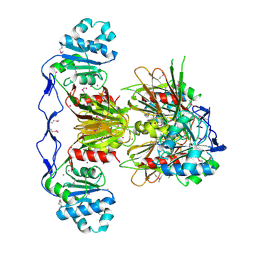 | | Crystal Structure of human NAD kinase | | Descriptor: | NAD kinase, UNKNOWN ATOM OR ION | | Authors: | Wang, H, Tempel, W, Wernimont, A.K, Tong, Y, Guan, X, Shen, Y, Li, Y, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-28 | | Release date: | 2010-11-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of human NAD kinase
to be published
|
|
8X0U
 
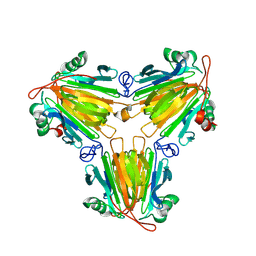 | |
8X0V
 
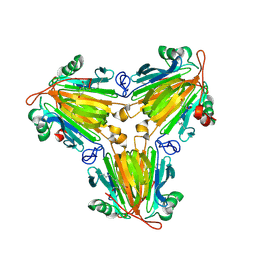 | |
5IZR
 
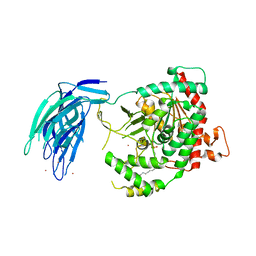 | | Human GIVD cytosolic phospholipase A2 in complex with Methyl gamma-Linolenyl Fluorophosphonate inhibitor and Terbium Chloride | | Descriptor: | Cytosolic phospholipase A2 delta, TERBIUM(III) ION, methyl (R)-(6Z,9Z,12Z)-octadeca-6,9,12-trien-1-ylphosphonofluoridate | | Authors: | Wang, H, Klein, M.G. | | Deposit date: | 2016-03-25 | | Release date: | 2016-06-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of Human GIVD Cytosolic Phospholipase A2 Reveals Insights into Substrate Recognition.
J.Mol.Biol., 428, 2016
|
|
5IZ5
 
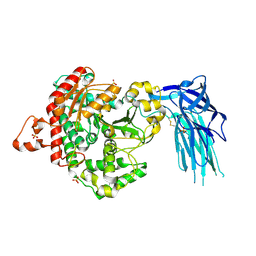 | | Human GIVD cytosolic phospholipase A2 | | Descriptor: | Cytosolic phospholipase A2 delta, SULFATE ION | | Authors: | Wang, H, Klein, M.G. | | Deposit date: | 2016-03-24 | | Release date: | 2016-06-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Human GIVD Cytosolic Phospholipase A2 Reveals Insights into Substrate Recognition.
J.Mol.Biol., 428, 2016
|
|
5IXC
 
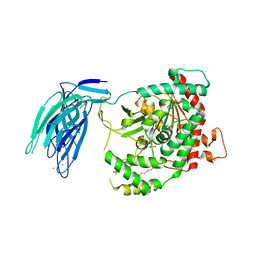 | | Human GIVD cytosolic phospholipase A2 in complex with Methyl gamma-Linolenyl Fluorophosphonate | | Descriptor: | BARIUM ION, Cytosolic phospholipase A2 delta, methyl (R)-(6Z,9Z,12Z)-octadeca-6,9,12-trien-1-ylphosphonofluoridate | | Authors: | Wang, H, Klein, M.G. | | Deposit date: | 2016-03-23 | | Release date: | 2016-06-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of Human GIVD Cytosolic Phospholipase A2 Reveals Insights into Substrate Recognition.
J.Mol.Biol., 428, 2016
|
|
8JIZ
 
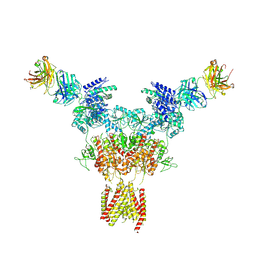 | |
8JJ2
 
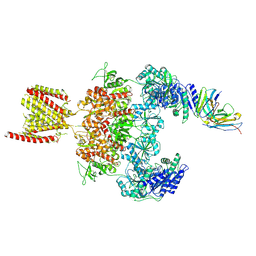 | | Cryo-EM structure of GluN1-2A NMDAR in complex with human Fab2G7 in one fab conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab2G7 Heavy Chain, ... | | Authors: | Wang, H, Zhu, S. | | Deposit date: | 2023-05-29 | | Release date: | 2024-06-05 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for antibody-mediated NMDA receptor clustering and endocytosis in autoimmune encephalitis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8JJ1
 
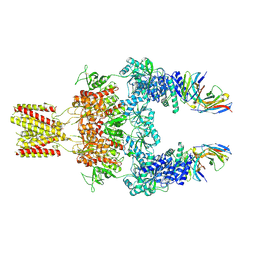 | | Cryo-EM structure of GluN1-2A NMDAR in complex with human Fab2G7 in two fab conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 2G7 Heavy Chain, ... | | Authors: | Wang, H, Zhu, S. | | Deposit date: | 2023-05-29 | | Release date: | 2024-06-05 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis for antibody-mediated NMDA receptor clustering and endocytosis in autoimmune encephalitis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8JJ0
 
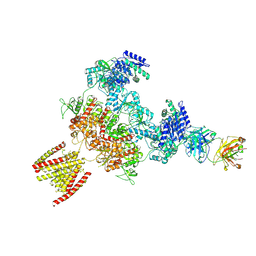 | |
3V93
 
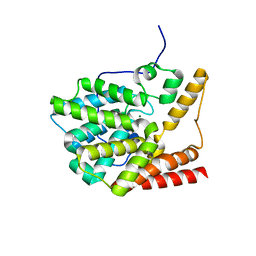 | | unliganded structure of TcrPDEC1 catalytic domain | | Descriptor: | Cyclic nucleotide specific phosphodiesterase, MAGNESIUM ION, ZINC ION | | Authors: | Wang, H, Kunz, S, Chen, G, Seebeck, T, Wan, Y, Robinson, H, Martinelli, S, Ke, H. | | Deposit date: | 2011-12-23 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biological and structural characterization of Trypanosoma cruzi phosphodiesterase C and Implications for design of parasite selective inhibitors.
J.Biol.Chem., 287, 2012
|
|
9COP
 
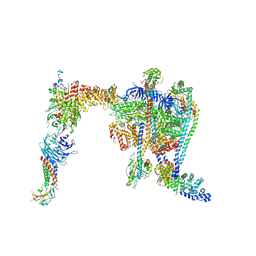 | | Yeast RAVE bound to V-ATPase V1 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Regulator of V-ATPase in vacuolar membrane protein 1, ... | | Authors: | Wang, H, Rubinstein, J.L. | | Deposit date: | 2024-07-17 | | Release date: | 2024-11-20 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of yeast RAVE bound to a partial V 1 complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
3V94
 
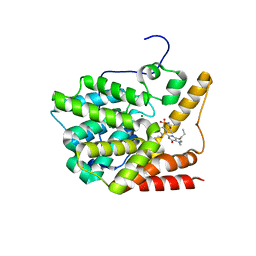 | | TcrPDEC1 catalytic domain in complex with inhibitor wyq16 | | Descriptor: | Cyclic nucleotide specific phosphodiesterase, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wang, H, Kunz, S, Chen, G, Seebeck, T, Wan, Y, Robinson, H, Martinelli, S, Ke, H. | | Deposit date: | 2011-12-23 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | TcrPDEC1 catalytic domain in complex with inhibitor wyq16
To be Published
|
|
6CM3
 
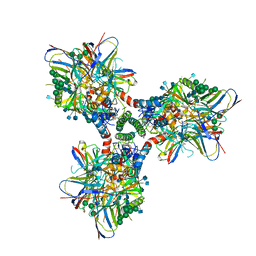 | | BG505 SOSIP in complex with sCD4, 17b, 8ANC195 | | Descriptor: | 17b Fab heavy chain, 17b Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, H, Bjorkman, P.J. | | Deposit date: | 2018-03-02 | | Release date: | 2018-10-17 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Partially Open HIV-1 Envelope Structures Exhibit Conformational Changes Relevant for Coreceptor Binding and Fusion.
Cell Host Microbe, 24, 2018
|
|
5F18
 
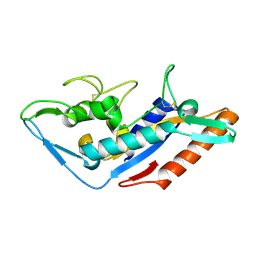 | | Structural basis of Ebola virus entry: viral glycoprotein bound to its endosomal receptor Niemann-Pick C1 | | Descriptor: | Niemann-Pick C1 protein | | Authors: | Wang, H, Shi, Y, Song, J, Qi, J, Lu, G, Yan, J, Gao, G.F. | | Deposit date: | 2015-11-30 | | Release date: | 2016-01-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ebola Viral Glycoprotein Bound to Its Endosomal Receptor Niemann-Pick C1.
Cell, 164, 2016
|
|
5F1B
 
 | | Structural basis of Ebola virus entry: viral glycoprotein bound to its endosomal receptor Niemann-Pick C1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GP1, GP2, ... | | Authors: | Wang, H, Shi, Y, Song, J, Qi, J, Lu, G, Yan, J, Gao, G.F. | | Deposit date: | 2015-11-30 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ebola Viral Glycoprotein Bound to Its Endosomal Receptor Niemann-Pick C1.
Cell, 164, 2016
|
|
8BZN
 
 | | SARS-CoV-2 non-structural protein 10 (nsp10) variant T102I | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Replicase polyprotein 1ab, ... | | Authors: | Wang, H, Rizvi, S.R.A, Dong, D, Lou, J, Wang, Q, Sopipong, W, Najar, F, Agarwal, P.K, Kozielski, F, Haider, S. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Emerging variants of SARS-CoV-2 NSP10 highlight strong functional conservation of its binding to two non-structural proteins, NSP14 and NSP16.
Elife, 12, 2023
|
|
6BYF
 
 | |
5THR
 
 | | Cryo-EM structure of a BG505 Env-sCD4-17b-8ANC195 complex | | Descriptor: | 17b Fab VH domain, 17b Fab VL domain, 8ANC195 G52K5 VH domain, ... | | Authors: | Wang, H, Bjorkman, P.J. | | Deposit date: | 2016-09-30 | | Release date: | 2016-11-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Cryo-EM structure of a CD4-bound open HIV-1 envelope trimer reveals structural rearrangements of the gp120 V1V2 loop.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3WOH
 
 | |
6LNY
 
 | |
2ZUL
 
 | | Crystal structure of Thermus thermophilus 16S rRNA methyltransferase RsmC (TTHA0533) in complex with cofactor S-adenosyl-L-Methionine | | Descriptor: | Probable ribosomal RNA small subunit methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Wang, H, Kawazoe, M, Tatsuguchi, A, Naoe, C, Takemoto, C, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-10-21 | | Release date: | 2009-10-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Thermus thermophilus 16S rRNA methyltransferase RsmC (TTHA0533) in complex with cofactor S-adenosyl-L-Methionine
To be Published
|
|
4O4D
 
 | | Crystal Structure of an Inositol hexakisphosphate kinase EhIP6KA in complexed with ATP and Ins(1,4,5)P3 | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2013-12-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | IP6K structure and the molecular determinants of catalytic specificity in an inositol phosphate kinase family.
Nat Commun, 5, 2014
|
|
