3HPN
 
 | | Ligand recognition by A-class EPH receptors: crystal structures of the EPHA2 ligand-binding domain and the EPHA2/EPHRIN-A1 complex | | Descriptor: | Ephrin type-A receptor 2 | | Authors: | Himanen, J.P, Goldgur, Y, Miao, H, Myshkin, E, Guo, H, Buck, M, Nguyen, M, Rajashankar, K.R, Wang, B, Nikolov, D.B. | | Deposit date: | 2009-06-04 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Ligand recognition by A-class Eph receptors: crystal structures of the EphA2 ligand-binding domain and the EphA2/ephrin-A1 complex.
Embo Rep., 10, 2009
|
|
8C41
 
 | | High resolution structure of the Streptococcus pneumoniae topoisomerase IV-DNA complex with the novel fluoroquinolone Delafloxacin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, DNA (5'-D(*CP*AP*TP*GP*AP*AP*T)-3'), ... | | Authors: | Najmudin, S, Pan, X.S, Wang, B, Chayen, N.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2022-12-30 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | The nature of the molecular interactions at high resolution of the Streptococcus pneumoniae topoisomerase IV-DNA complex with the novel fluoroquinolone Delafloxacin.
To Be Published
|
|
5EPO
 
 | | The three-dimensional structure of Clostridium absonum 7alpha-hydroxysteroid dehydrogenase | | Descriptor: | 7-alpha-hydroxysteroid deydrogenase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lou, D, Wang, B, Wang, F. | | Deposit date: | 2015-11-12 | | Release date: | 2016-03-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The three-dimensional structure of Clostridium absonum 7 alpha-hydroxysteroid dehydrogenase: new insights into the conserved arginines for NADP(H) recognition
Sci Rep, 6, 2016
|
|
2YXM
 
 | | Crystal structure of I-set domain of human Myosin Binding ProteinC | | Descriptor: | Myosin-binding protein C, slow-type | | Authors: | Kishishita, S, Ohsawa, N, Murayama, K, Chen, L, Liu, Z, Terada, T, Shirouzu, M, Wang, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-26 | | Release date: | 2007-10-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of I-set domain of human Myosin Binding ProteinC
To be Published
|
|
7DCN
 
 | | Apo-citrate lyase phosphoribosyl-dephospho-CoA transferase | | Descriptor: | Probable apo-citrate lyase phosphoribosyl-dephospho-CoA transferase, SULFATE ION, ZINC ION | | Authors: | Xu, H, Wang, B, Su, X.D. | | Deposit date: | 2020-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Co-evolution-based prediction of metal-binding sites in proteomes by machine learning.
Nat.Chem.Biol., 19, 2023
|
|
7DCM
 
 | | Crystal structure of CITX | | Descriptor: | Probable apo-citrate lyase phosphoribosyl-dephospho-CoA transferase, ZINC ION | | Authors: | Xu, H, Wang, B, Su, X.D. | | Deposit date: | 2020-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Co-evolution-based prediction of metal-binding sites in proteomes by machine learning.
Nat.Chem.Biol., 19, 2023
|
|
6VKJ
 
 | |
1O06
 
 | | Crystal structure of the Vps27p Ubiquitin Interacting Motif (UIM) | | Descriptor: | Vacuolar protein sorting-associated protein VPS27, ZINC ION | | Authors: | Fisher, R.D, Wang, B, Alam, S.L, Higginson, D.S, Rich, R, Myszka, D, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2003-02-20 | | Release date: | 2003-07-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and ubiquitin binding of the ubiquitin-interacting motif.
J.Biol.Chem., 278, 2003
|
|
6LKT
 
 | | Crystal structure of the Fab fragment of murine monoclonal antibody KH-1 against Human herpesvirus 6B | | Descriptor: | antibody Fab Fragment L-chain, antibody Fab fragment H chain | | Authors: | Nishimura, M, Novita, B.D, Kato, T, Tjan, L.H, Wang, B, Wakata, A, Poetranto, A.L, Kawabata, A, Tang, H, Aoshi, T, Mori, Y. | | Deposit date: | 2019-12-20 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the interaction of human herpesvirus 6B tetrameric glycoprotein complex with the cellular receptor, human CD134.
Plos Pathog., 16, 2020
|
|
8HB9
 
 | | Crystal Structure of Human IDH1 R132H Mutant in Complex with NADPH and Compound IHMT-IDH1-053 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | Authors: | Guo, G, Wang, B, Liu, J, Liu, Q. | | Deposit date: | 2022-10-27 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based discovery of IHMT-IDH1-053 as a potent irreversible IDH1 mutant selective inhibitor.
Eur.J.Med.Chem., 256, 2023
|
|
6LTG
 
 | | Crystal structure of the Fab fragment of murine monoclonal antibody OHV-3 against Human herpesvirus 6B | | Descriptor: | MAGNESIUM ION, antibody Fab fragment H-chain, antibody Fab fragment L-chain | | Authors: | Nishimura, M, Novita, B.D, Kato, T, Tjan, L.H, Wang, B, Wakata, A, Poetranto, A.L, Kawabata, A, Tang, H, Aoshi, T, Mori, Y. | | Deposit date: | 2020-01-22 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis for the interaction of human herpesvirus 6B tetrameric glycoprotein complex with the cellular receptor, human CD134.
Plos Pathog., 16, 2020
|
|
5WWN
 
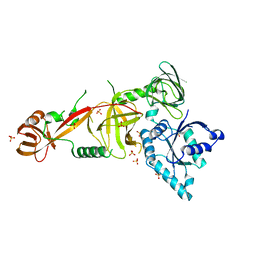 | | Crystal structure of Tsr1 | | Descriptor: | Ribosome biogenesis protein TSR1, SULFATE ION | | Authors: | Ye, K, Wang, B. | | Deposit date: | 2017-01-03 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Molecular architecture of the 90S small subunit pre-ribosome
Elife, 6, 2017
|
|
1YSN
 
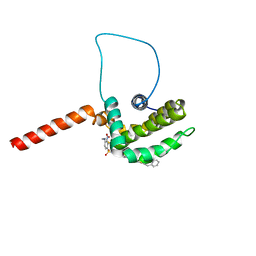 | | Solution structure of the anti-apoptotic protein Bcl-xL complexed with an acyl-sulfonamide-based ligand | | Descriptor: | 3-NITRO-N-{4-[2-(2-PHENYLETHYL)-1,3-BENZOTHIAZOL-5-YL]BENZOYL}-4-{[2-(PHENYLSULFANYL)ETHYL]AMINO}BENZENESULFONAMIDE, Apoptosis regulator Bcl-X | | Authors: | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | Deposit date: | 2005-02-08 | | Release date: | 2005-06-07 | | Last modified: | 2023-11-29 | | Method: | SOLUTION NMR | | Cite: | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
1YSI
 
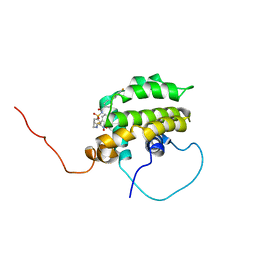 | | Solution structure of the anti-apoptotic protein Bcl-xL in complex with an acyl-sulfonamide-based ligand | | Descriptor: | Apoptosis regulator Bcl-X, N-[(4'-FLUORO-1,1'-BIPHENYL-4-YL)CARBONYL]-3-NITRO-4-{[2-(PHENYLSULFANYL)ETHYL]AMINO}BENZENESULFONAMIDE | | Authors: | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | Deposit date: | 2005-02-08 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
1YSX
 
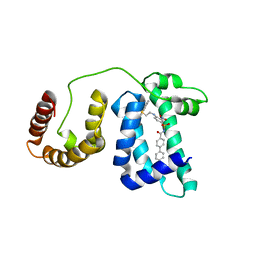 | | Solution structure of domain 3 from human serum albumin complexed to an anti-apoptotic ligand directed against Bcl-xL and Bcl-2 | | Descriptor: | 4-({2-[(2,4-DIMETHYLPHENYL)SULFANYL]ETHYL}AMINO)-N-[(4'-FLUORO-1,1'-BIPHENYL-4-YL)CARBONYL]-3-NITROBENZENESULFONAMIDE, Serum albumin | | Authors: | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | Deposit date: | 2005-02-09 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
5NSD
 
 | | Co-crystal structure of NAMPT dimer with KPT-9274 | | Descriptor: | (~{E})-3-(6-azanylpyridin-3-yl)-~{N}-[[5-[4-[4,4-bis(fluoranyl)piperidin-1-yl]carbonylphenyl]-7-(4-fluorophenyl)-1-benzofuran-2-yl]methyl]prop-2-enamide, GLYCEROL, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Neggers, J.E, Kwanten, B, Dierckx, T, Noguchi, H, Voet, A, Vercruysse, T, Baloglu, E, Senapedis, W, Jacquemyn, M, Daelemans, D. | | Deposit date: | 2017-04-26 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.046 Å) | | Cite: | Target identification of small molecules using large-scale CRISPR-Cas mutagenesis scanning of essential genes.
Nat Commun, 9, 2018
|
|
5ANM
 
 | | Crystal structure of IgE Fc in complex with a neutralizing antibody | | Descriptor: | IG EPSILON CHAIN C REGION, IMMUNOGLOBULIN G, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cohen, E.S, Dobson, C.L, Kack, H, Wang, B, Sims, D.A, Lloyd, C.O, England, E, Rees, D.G, Guo, H, Karagiannis, S.N, O'Brien, S, Persdotter, S, Ekdahl, H, Butler, R, Keyes, F, Oakley, S, Carlsson, M, Briend, E, Wilkinson, T, Anderson, I.K, Monk, P.D, vonWachenfeldt, K, Eriksson, P.O, Gould, H.J, Vaughan, T.J, May, R.D. | | Deposit date: | 2015-09-07 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A Novel Ige-Neutralizing Antibody for the Treatment of Severe Uncontrolled Asthma.
Mabs, 6, 2015
|
|
1YSG
 
 | | Solution Structure of the Anti-apoptotic Protein Bcl-xL in Complex with "SAR by NMR" Ligands | | Descriptor: | 4'-FLUORO-1,1'-BIPHENYL-4-CARBOXYLIC ACID, 5,6,7,8-TETRAHYDRONAPHTHALEN-1-OL, Apoptosis regulator Bcl-X | | Authors: | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | Deposit date: | 2005-02-08 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
1YSW
 
 | | Solution structure of the anti-apoptotic protein Bcl-2 complexed with an acyl-sulfonamide-based ligand | | Descriptor: | 3-NITRO-N-{4-[2-(2-PHENYLETHYL)-1,3-BENZOTHIAZOL-5-YL]BENZOYL}-4-{[2-(PHENYLSULFANYL)ETHYL]AMINO}BENZENESULFONAMIDE, Apoptosis regulator Bcl-2 | | Authors: | Oltersdorf, T, Elmore, S.W, Shoemaker, A.R, Armstrong, R.C, Augeri, D.J, Belli, B.A, Bruncko, M, Deckwerth, T.L, Dinges, J, Hajduk, P.J, Joseph, M.K, Kitada, S, Korsmeyer, S.J, Kunzer, A.R, Letai, A, Li, C, Mitten, M.J, Nettesheim, D.G, Ng, S, Nimmer, P.M, O'Connor, J.M, Oleksijew, A, Petros, A.M, Reed, J.C, Shen, W, Tahir, S.K, Thompson, C.B, Tomaselli, K.J, Wang, B, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H. | | Deposit date: | 2005-02-09 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An inhibitor of Bcl-2 family proteins induces regression of solid tumours
Nature, 435, 2005
|
|
1ZGH
 
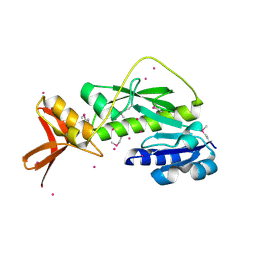 | | Methionyl-tRNA formyltransferase from Clostridium thermocellum | | Descriptor: | Methionyl-tRNA formyltransferase, UNKNOWN ATOM OR ION | | Authors: | Yang, H, Kataeva, I, Xu, H, Zhao, M, Chang, J, Liu, Z, Chen, L, Tempel, W, Habel, J, Zhou, W, Lee, D, Lin, D, Chang, S, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Rose, J.P, Wang, B, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-04-21 | | Release date: | 2005-05-03 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Methionyl-tRNA formyltransferase from Clostridium thermocellum
To be published
|
|
7JSJ
 
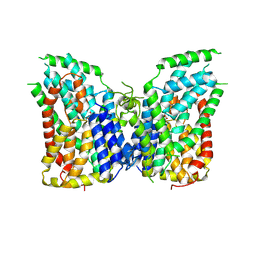 | | Structure of the NaCT-PF2 complex | | Descriptor: | (2R)-2-[2-(4-tert-butylphenyl)ethyl]-2-hydroxybutanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, ... | | Authors: | Sauer, D.B, Wang, B, Song, J, Rice, W.J, Wang, D.N. | | Deposit date: | 2020-08-14 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structure and inhibition mechanism of the human citrate transporter NaCT.
Nature, 591, 2021
|
|
7JSK
 
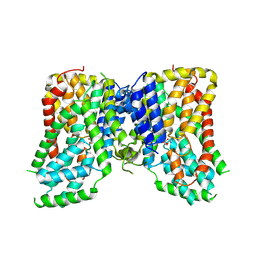 | | Structure of the NaCT-Citrate complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, SODIUM ION, ... | | Authors: | Sauer, D.B, Wang, B, Song, J, Rice, W.J, Wang, D.N. | | Deposit date: | 2020-08-14 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structure and inhibition mechanism of the human citrate transporter NaCT.
Nature, 591, 2021
|
|
6J4F
 
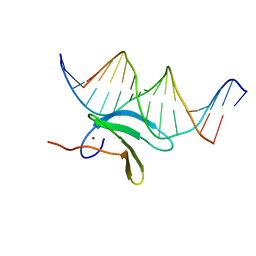 | | Crystal structure of the AtWRKY2 domain | | Descriptor: | DNA (5'-D(*AP*GP*CP*CP*TP*TP*TP*GP*AP*CP*CP*AP*GP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*CP*TP*GP*GP*TP*CP*AP*AP*AP*GP*GP*C)-3'), Probable WRKY transcription factor 2, ... | | Authors: | Xu, Y.P, Xu, H, Wang, B, Su, X.D. | | Deposit date: | 2019-01-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the AtWRKY2 domain
To Be Published
|
|
4HY1
 
 | | Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. | | Descriptor: | 6-({4-[(3R)-3-aminopyrrolidin-1-yl]-5-chloro-6-ethyl-7H-pyrrolo[2,3-d]pyrimidin-2-yl}sulfanyl)-1H-isoindol-1-one, Topoisomerase IV, subunit B | | Authors: | Bensen, D.C, Creighton, C.J, Kwan, B, Tari, L.W. | | Deposit date: | 2012-11-12 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6J4G
 
 | | Crystal structure of the AtWRKY33 domain | | Descriptor: | DNA (5'-D(*AP*GP*CP*CP*TP*TP*TP*GP*AP*CP*CP*AP*GP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*CP*TP*GP*GP*TP*CP*AP*AP*AP*GP*GP*C)-3'), Probable WRKY transcription factor 33, ... | | Authors: | Xu, Y.P, Xu, H, Wang, B, Su, X.D. | | Deposit date: | 2019-01-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the N-terminal DNA binding domain of AtWRKY33
To Be Published
|
|
