6LOQ
 
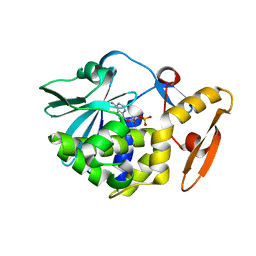 | | crystal structure of alpha-momorcharin in complex with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Ribosome-inactivating protein momordin I | | Authors: | Fan, X, Jin, T. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.331 Å) | | Cite: | Atomic-resolution structures of type I ribosome inactivating protein alpha-momorcharin with different substrate analogs.
Int.J.Biol.Macromol., 164, 2020
|
|
6LOY
 
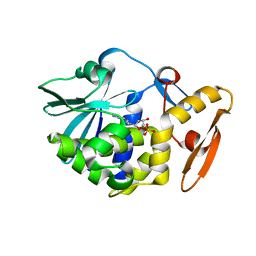 | | crystal structure of alpha-momorcharin in complex with dAMP | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Ribosome-inactivating protein momordin I | | Authors: | Fan, X, Jin, T. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Atomic-resolution structures of type I ribosome inactivating protein alpha-momorcharin with different substrate analogs.
Int.J.Biol.Macromol., 164, 2020
|
|
5WZX
 
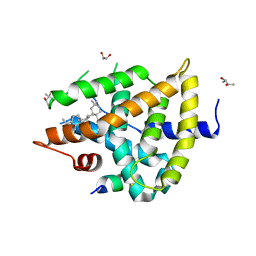 | | Structural basis for a pentacyclic oleanane-type triterpenoid as a ligand of FXR | | Descriptor: | (4aR,6aR,6aS,6bS,8aS,9R,12aR,14bR)-2,2,6a,6b,9,12a-hexamethyl-10-oxidanylidene-1,3,4,5,6,6a,7,8,8a,9,11,12,13,14b-tetradecahydropicene-4a-carboxylic acid, (R,R)-2,3-BUTANEDIOL, Bile acid receptor, ... | | Authors: | Lu, Y, Li, Y. | | Deposit date: | 2017-01-19 | | Release date: | 2018-01-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Identification of an Oleanane-Type Triterpene Hedragonic Acid as a Novel Farnesoid X Receptor Ligand with Liver Protective Effects and Anti-inflammatory Activity
Mol. Pharmacol., 93, 2018
|
|
5WPW
 
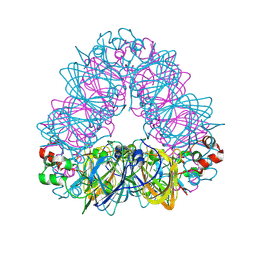 | | Crystal structure of coconut allergen cocosin | | Descriptor: | 11S globulin isoform 1 | | Authors: | Jin, T, Zhang, Y. | | Deposit date: | 2016-11-21 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Crystal Structure of Cocosin, A Potential Food Allergen from Coconut (Cocos nucifera)
J. Agric. Food Chem., 65, 2017
|
|
2HOI
 
 | |
2HOF
 
 | |
1KBU
 
 | | CRE RECOMBINASE BOUND TO A LOXP HOLLIDAY JUNCTION | | Descriptor: | CRE RECOMBINASE, LOXP | | Authors: | Martin, S.S, Pulido, E, Chu, V.C, Lechner, T, Baldwin, E.P. | | Deposit date: | 2001-11-06 | | Release date: | 2002-06-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Order of Strand Exchanges in Cre-LoxP Recombination and its Basis Suggested by the Crystal Structure of a
Cre-LoxP Holliday Junction Complex
J.Mol.Biol., 319, 2002
|
|
3FZ3
 
 | | Crystal Structure of almond Pru1 protein | | Descriptor: | CALCIUM ION, Prunin, SODIUM ION | | Authors: | Jin, T.C, Zhang, Y.Z. | | Deposit date: | 2009-01-23 | | Release date: | 2009-11-10 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of prunin-1, a major component of the almond (Prunus dulcis) allergen amandin.
J.Agric.Food Chem., 57, 2009
|
|
9JNN
 
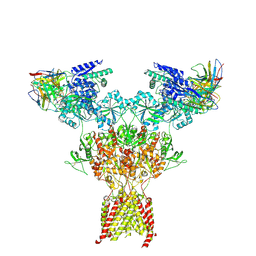 | | Structure of native di-heteromeric GluN1-GluN2B NMDA receptor in rat cortex and hippocampus | | Descriptor: | (2R)-4-(3-phosphonopropyl)piperazine-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, M, Feng, J, Li, Y, Zhu, S. | | Deposit date: | 2024-09-23 | | Release date: | 2025-02-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Assembly and architecture of endogenous NMDA receptors in adult cerebral cortex and hippocampus.
Cell, 188, 2025
|
|
1MA7
 
 | |
3DBO
 
 | | Crystal structure of a member of the VapBC family of toxin-antitoxin systems, VapBC-5, from Mycobacterium tuberculosis | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, SODIUM ION, ... | | Authors: | Miallau, L, Cascio, D, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2008-06-02 | | Release date: | 2008-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure and Proposed Activity of a Member of the VapBC Family of Toxin-Antitoxin Systems: VapBC-5 FROM MYCOBACTERIUM TUBERCULOSIS.
J.Biol.Chem., 284, 2009
|
|
3C3V
 
 | |
4ZRI
 
 | | Crystal structure of Merlin-FERM and Lats2 | | Descriptor: | Merlin, Serine/threonine-protein kinase LATS2 | | Authors: | Li, F, Zhou, H, Long, J, Shen, Y. | | Deposit date: | 2015-05-12 | | Release date: | 2015-06-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway
Cell Res., 25, 2015
|
|
4ZRJ
 
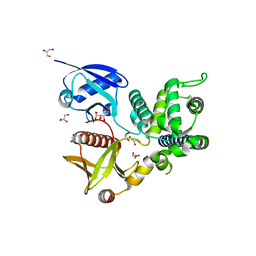 | | Structure of Merlin-FERM and CTD | | Descriptor: | GLYCEROL, Merlin | | Authors: | Lin, Z, Li, F, Long, J, Shen, Y. | | Deposit date: | 2015-05-12 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway
Cell Res., 25, 2015
|
|
4ZRK
 
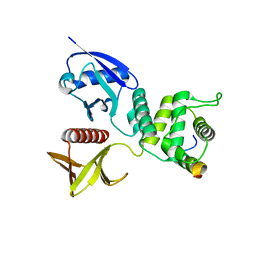 | | Merlin-FERM and Lats1 complex | | Descriptor: | Merlin, Serine/threonine-protein kinase LATS1 | | Authors: | Lin, Z, Li, Y, Wei, Z, Zhang, M. | | Deposit date: | 2015-05-12 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.316 Å) | | Cite: | Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway
Cell Res., 25, 2015
|
|
7YFM
 
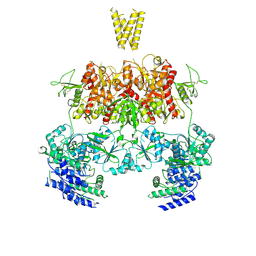 | | Structure of GluN1b-GluN2D NMDA receptor in complex with agonists glycine and glutamate. | | Descriptor: | Glutamate receptor ionotropic, NMDA 2D, Isoform 6 of Glutamate receptor ionotropic, ... | | Authors: | Zhang, J.L, Zhu, S.J, Zhang, M. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFF
 
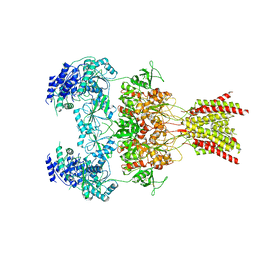 | | Structure of GluN1a-GluN2D NMDA receptor in complex with agonist glycine and competitive antagonist CPP. | | Descriptor: | (2R)-4-(3-phosphonopropyl)piperazine-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J.L, Zhu, S.J, Zhang, M. | | Deposit date: | 2022-07-08 | | Release date: | 2023-04-12 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFL
 
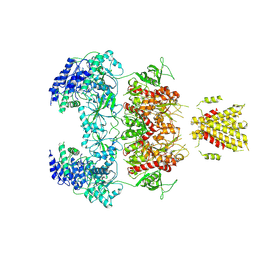 | | Structure of GluN1a-GluN2D NMDA receptor in complex with agonists glycine and glutamate. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, J.L, Zhu, S.J, Zhang, M. | | Deposit date: | 2022-07-08 | | Release date: | 2023-04-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFO
 
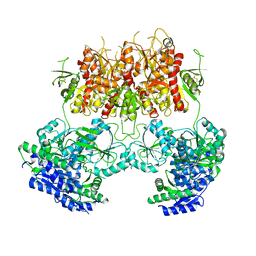 | |
7YFR
 
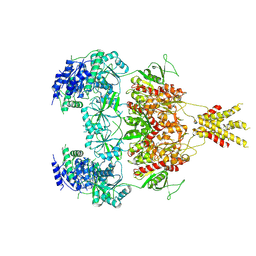 | | Structure of GluN1a E698C-GluN2D NMDA receptor in cystines non-crosslinked state. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, J.L, Zhu, S.J, Zhang, M. | | Deposit date: | 2022-07-09 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7XJ2
 
 | | Structure of human TRPV3_G573S in complex with Trpvicin in C4 symmetry | | Descriptor: | Fusion protein of Transient receptor potential cation channel subfamily V member 3 and 3C-GFP, N-[5-[2-(2-cyanopropan-2-yl)pyridin-4-yl]-4-(trifluoromethyl)-1,3-thiazol-2-yl]-4,6-dimethoxy-pyrimidine-5-carboxamide | | Authors: | Fan, J, Yue, Z, Jiang, D, Lei, X. | | Deposit date: | 2022-04-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
7XJ1
 
 | | Structure of human TRPV3_G573S in complex with Trpvicin in C2 symmetry | | Descriptor: | Fusion protein of Transient receptor potential cation channel subfamily V member 3 and 3C-GFP, N-[5-[2-(2-cyanopropan-2-yl)pyridin-4-yl]-4-(trifluoromethyl)-1,3-thiazol-2-yl]-4,6-dimethoxy-pyrimidine-5-carboxamide, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Fan, J, Yue, Z, Jiang, D, Lei, X. | | Deposit date: | 2022-04-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
7XJ3
 
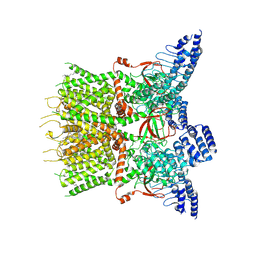 | | Structure of human TRPV3 | | Descriptor: | [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate, fusion of transient receptor potential cation channel subfamily V member 3 and 3C-GFP | | Authors: | Fan, J, Yue, Z, Jiang, D, Lei, X. | | Deposit date: | 2022-04-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
7XJ0
 
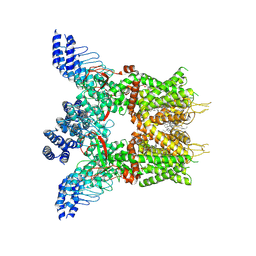 | | Structure of human TRPV3 in complex with Trpvicin | | Descriptor: | Fusion protein of Transient receptor potential cation channel subfamily V member 3 and 3C-GFP, N-[5-[2-(2-cyanopropan-2-yl)pyridin-4-yl]-4-(trifluoromethyl)-1,3-thiazol-2-yl]-4,6-dimethoxy-pyrimidine-5-carboxamide, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Fan, J, Yue, Z, Jiang, D, Lei, X. | | Deposit date: | 2022-04-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
7Y5N
 
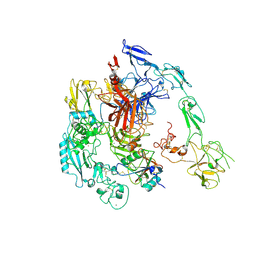 | | Structure of 1:1 PAPP-A.ProMBP complex(half map) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bone marrow proteoglycan, ... | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-06-17 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
