5NDH
 
 | | The structure of the G. violaceus guanidine II riboswitch P2 stem-loop | | Descriptor: | GUANIDINE, MAGNESIUM ION, RNA (5'-R(*GP*(CBV)P*GP*GP*GP*GP*AP*CP*GP*AP*CP*CP*CP*CP*GP*C)-3'), ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-03-08 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The Structure of the Guanidine-II Riboswitch.
Cell Chem Biol, 24, 2017
|
|
3VAC
 
 | |
8BFW
 
 | | The structures of Ace2 in complex with bicyclic peptide inhibitor | | Descriptor: | 1-[3,5-bis(3-bromanylpropanoyl)-1,3,5-triazinan-1-yl]-3-bromanyl-propan-1-one, ALA-CYS-VAL-ARG-SER-HIS-CYS-SER-SER-LEU-LEU-PRO-ARG-ILE-HIS-CYS-ALA-NH2, Processed angiotensin-converting enzyme 2 | | Authors: | Brear, P, Lulla, A, Harman, M, Dods, R, Chen, L, Bezerra, G, Demydchuk, Y, Stanway, S, Hyvonen, M. | | Deposit date: | 2022-10-27 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure-Guided Chemical Optimization of Bicyclic Peptide ( Bicycle ) Inhibitors of Angiotensin-Converting Enzyme 2.
J.Med.Chem., 66, 2023
|
|
5NDX
 
 | | The bacterial orthologue of Human a-L-iduronidase does not need N-glycan post-translational modifications to be catalytically competent: Crystallography and QM/MM insights into Mucopolysaccharidosis I | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-6-(4-methyl-2-oxidanylidene-chromen-7-yl)oxy-3,4,5-tris(oxidanyl)oxane-2-carboxylic acid, Glycosyl hydrolase, SULFATE ION | | Authors: | Raich, L, Valero-Gonzalez, J, Castro-Lopez, J, Millan, C, Jimenez-Garcia, M.J, Nieto, P, Uson, I, Hurtado-Guerrero, R, Rovira, C. | | Deposit date: | 2017-03-09 | | Release date: | 2018-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The bacterial orthologue of Human a-L-iduronidase does not need N-glycan post-translational modifications to be catalytically competent:
Crystallography and QM/MM insights into Mucopolysaccharidosis I.
To Be Published
|
|
5N6H
 
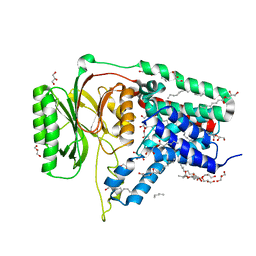 | | Structure of the membrane integral lipoprotein N-acyltransferase Lnt from E. coli | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apolipoprotein N-acyltransferase, GLYCEROL | | Authors: | Huang, C.-Y, Boland, C, Howe, N, Wiktor, M, Vogeley, L, Weichert, D, Bailey, J, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2017-02-15 | | Release date: | 2017-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the mechanism of the membrane integral N-acyltransferase step in bacterial lipoprotein synthesis.
Nat Commun, 8, 2017
|
|
5YMR
 
 | | The Crystal Structure of IseG | | Descriptor: | 2-hydroxyethylsulfonic acid, Formate acetyltransferase, GLYCEROL | | Authors: | Lin, L, Zhang, J, Xing, M, Hua, G, Guo, C, Hu, Y, Wei, Y, Ang, E, Zhao, H, Zhang, Y, Yuchi, Z. | | Deposit date: | 2017-10-22 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Radical-mediated C-S bond cleavage in C2 sulfonate degradation by anaerobic bacteria.
Nat Commun, 10, 2019
|
|
5MZ6
 
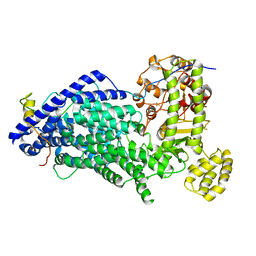 | | Cryo-EM structure of a Separase-Securin complex from Caenorhabditis elegans at 3.8 A resolution | | Descriptor: | Interactor of FizzY protein, SEParase | | Authors: | Boland, A, Martin, T.G, Zhang, Z, Yang, J, Bai, X.C, Chang, L, Scheres, S.H.W, Barford, D. | | Deposit date: | 2017-01-31 | | Release date: | 2017-03-08 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of a metazoan separase-securin complex at near-atomic resolution.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5YNP
 
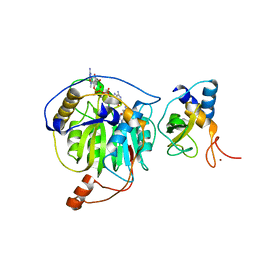 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to sinefungin and m7GpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, SINEFUNGIN, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-25 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YPA
 
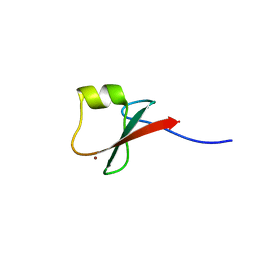 | | p62/SQSTM1 ZZ domain with Lys-peptide | | Descriptor: | 78 kDa glucose-regulated protein,Sequestosome-1, ZINC ION | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-11-01 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter.
Nat Commun, 9, 2018
|
|
6GA3
 
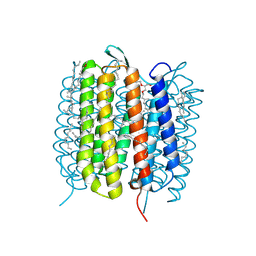 | | Bacteriorhodopsin, 33 ms state, ensemble refinement | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Barends, T.R.M, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
6GAC
 
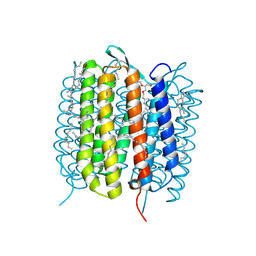 | | BACTERIORHODOPSIN, 490 FS STATE, REAL-SPACE REFINED AGAINST 15% EXTRAPOLATED STRUCTURE FACTORS | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
1DQ0
 
 | | Locked, metal-free concanavalin A, a minor species in solution | | Descriptor: | Concanavalin-Br | | Authors: | Bouckaert, J, Dewallef, Y, Poortmans, F, Wyns, L, Loris, R. | | Deposit date: | 1999-12-29 | | Release date: | 2000-01-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structural features of concanavalin A governing non-proline peptide isomerization
J.Biol.Chem., 275, 2000
|
|
4AC3
 
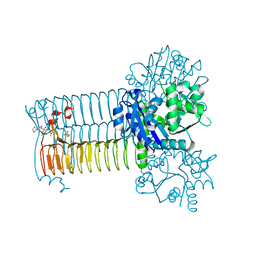 | | S.pneumoniae GlmU in complex with an antibacterial inhibitor | | Descriptor: | BIFUNCTIONAL PROTEIN GLMU, N-{2,4-DIMETHOXY-5-[(2-PIPERIDIN-1-YLBENZYL)sulfamoyl]phenyl}acetamide, SULFATE ION | | Authors: | Otterbein, L, Breed, J, Ogg, D.J. | | Deposit date: | 2011-12-12 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibitors of Acetyltransferase Domain of N-Acetylglucosamine-1-Phosphate-Uridyltransferase/ Glucosamine-1-Phosphate-Acetyltransferase (Glmu). Part 1: Hit to Lead Evaluation of a Novel Arylsulfonamide Series.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6G6P
 
 | | Crystal structure of the computationally designed Ika8 protein: crystal packing No.2 in P63 | | Descriptor: | Ika8 | | Authors: | Noguchi, H, Addy, C, Simoncini, D, Van Meervelt, L, Schiex, T, Zhang, K.Y.J, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2018-04-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Computational design of symmetrical eight-bladed beta-propeller proteins.
IUCrJ, 6, 2019
|
|
5YP7
 
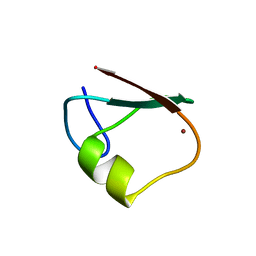 | | p62/SQSTM1 ZZ domain | | Descriptor: | Sequestosome-1, ZINC ION | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-11-01 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.424 Å) | | Cite: | Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter.
Nat Commun, 9, 2018
|
|
6GCU
 
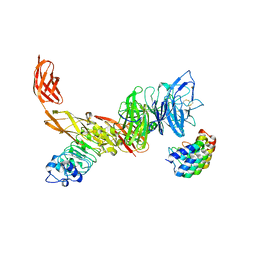 | | MET receptor in complex with InlB internalin domain and DARPin A3A | | Descriptor: | DARPin A3A, Hepatocyte growth factor receptor, Internalin B | | Authors: | Meyer, T, Andres, F, Iamele, L, Gherardi, E, Pluckthun, A, Niemann, H.H. | | Deposit date: | 2018-04-19 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (6.001 Å) | | Cite: | Inhibition of the MET Kinase Activity and Cell Growth in MET-Addicted Cancer Cells by Bi-Paratopic Linking.
J.Mol.Biol., 431, 2019
|
|
1E3U
 
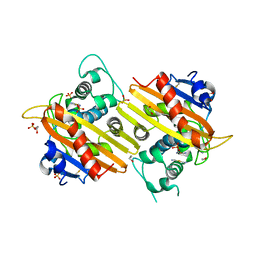 | | MAD structure of OXA10 class D beta-lactamase | | Descriptor: | 1,2-ETHANEDIOL, BETA-LACTAMASE OXA-10, GOLD (I) CYANIDE ION, ... | | Authors: | Maveyraud, L, Golemi, D, Kotra, L.P, Tranier, S, Vakulenko, S, Mobashery, S, Samama, J.P. | | Deposit date: | 2000-06-23 | | Release date: | 2001-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Insights Into Class D Beta-Lactamases are Revealed by the Crystal Structure of the Oxa10 Enzyme from Pseudomonas Aeruginosa
Structure, 8, 2000
|
|
8SJ3
 
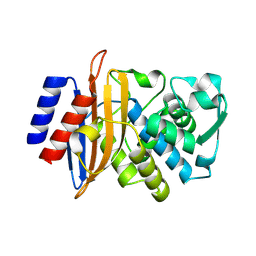 | | Beta-lactamase CTX-M-14 E166Y/N170G | | Descriptor: | Beta-lactamase CTX-M-14 | | Authors: | Judge, A, Palzkill, T, Sankaran, B, Prasad, B.V.V, Hu, L. | | Deposit date: | 2023-04-17 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Network of epistatic interactions in an enzyme active site revealed by large-scale deep mutational scanning.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5N05
 
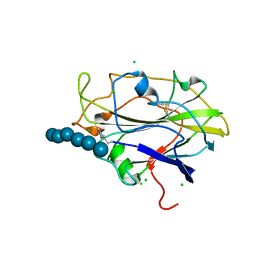 | | X-ray crystal structure of an LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, CHLORIDE ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.-C.N, Lo Leggio, L. | | Deposit date: | 2017-02-02 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Unliganded and substrate bound structures of the cellooligosaccharide active lytic polysaccharide monooxygenase LsAA9A at low pH.
Carbohydr. Res., 448, 2017
|
|
8STL
 
 | | Crystal Structure of Nanobody PIK3_Nb16 against wild-type PI3Kalpha | | Descriptor: | Nanobody PIK3_Nb16, SULFATE ION | | Authors: | Nwafor, J.N, Srinivasan, L, Chen, Z, Gabelli, S.B, Iheanacho, A, Alzogaray, V, Klinke, S. | | Deposit date: | 2023-05-10 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of isoform specific nanobodies for Class I PI3Ks
To be published
|
|
1DQ4
 
 | | A transient unlocked concanavalin A structure with MN2+ bound in the transition metal ion binding site S1 and an empty calcium binding site S2 | | Descriptor: | Concanavalin-Br, MANGANESE (II) ION | | Authors: | Bouckaert, J, Dewallef, Y, Poortmans, F, Wyns, L, Loris, R. | | Deposit date: | 1999-12-30 | | Release date: | 2000-01-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structural features of concanavalin A governing non-proline peptide isomerization
J.Biol.Chem., 275, 2000
|
|
6G89
 
 | | Thaumatin solved by Native SAD from a dataset collected in 0.6 second with JUNGFRAU detector | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Leonarski, F, Olieric, V, Vera, L, Redford, S, Wang, M. | | Deposit date: | 2018-04-08 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.359 Å) | | Cite: | Fast and accurate data collection for macromolecular crystallography using the JUNGFRAU detector.
Nat. Methods, 15, 2018
|
|
8SLI
 
 | | Cryo-EM structure of the rat TRPM5 channel in 2mM calcium, high-1 | | Descriptor: | CALCIUM ION, Transient receptor potential cation channel subfamily M member 5 | | Authors: | Karuppan, S, Schrag, L.G, Jara-Oseguera, A, Zubcevic, L. | | Deposit date: | 2023-04-21 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural dynamics at cytosolic interprotomer interfaces control gating of a mammalian TRPM5 channel.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
1NXK
 
 | | Crystal structure of staurosporine bound to MAP KAP kinase 2 | | Descriptor: | MAP kinase-activated protein kinase 2, STAUROSPORINE, SULFATE ION | | Authors: | Underwood, K.W, Parris, K.D, Federico, E, Mosyak, L, Czerwinski, R.M, Shane, T, Taylor, M, Svenson, K, Liu, Y, Hsiao, C.L, Wolfrom, S, Malakian, K, Telliez, J.B, Lin, L.L, Kriz, R.W, Seehra, J, Somers, W.S, Stahl, M.L. | | Deposit date: | 2003-02-10 | | Release date: | 2003-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Catalytically active MAP KAP kinase 2 structures in complex with staurosporine and ADP reveal differences with the autoinhibited enzyme
Structure, 11, 2003
|
|
5YLO
 
 | | Structural of Pseudomonas aeruginosa PA4980 | | Descriptor: | GLYCEROL, Probable enoyl-CoA hydratase/isomerase | | Authors: | Liu, L, Li, T, Peng, C.T, Li, C.C, Xiao, Q.J, He, L.H, Wang, N.Y, Bao, R. | | Deposit date: | 2017-10-18 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural characterization of a Delta3, Delta2-enoyl-CoA isomerase from Pseudomonas aeruginosa: implications for its involvement in unsaturated fatty acid metabolism.
J.Biomol.Struct.Dyn., 37, 2019
|
|
