4LZK
 
 | |
4KTB
 
 | | The crystal structure of posible asymmetric diadenosine tetraphosphate (Ap(4)A) hydrolases from Jonesia denitrificans DSM 20603 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Putative uncharacterized protein, ... | | Authors: | Tan, K, Kim, Y, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-20 | | Release date: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (1.936 Å) | | Cite: | The crystal structure of posible asymmetric diadenosine tetraphosphate (Ap(4)A) hydrolases from Jonesia denitrificans DSM 20603
To be Published
|
|
4KLK
 
 | | Phage-related protein DUF2815 from Enterococcus faecalis | | Descriptor: | ETHANOL, GLYCEROL, Phage-related protein DUF2815 | | Authors: | Osipiuk, J, Wu, R, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-07 | | Release date: | 2013-05-22 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Phage-related protein DUF2815 from Enterococcus faecalis
To be Published
|
|
4KND
 
 | | Thioredoxin from Anaeromyxobacter dehalogenans. | | Descriptor: | Thioredoxin | | Authors: | Osipiuk, J, Hatzos-Skintges, C, Clancy, S, Adkins, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2013-05-09 | | Release date: | 2013-05-22 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thioredoxin from Anaeromyxobacter dehalogenans.
To be Published
|
|
4KD5
 
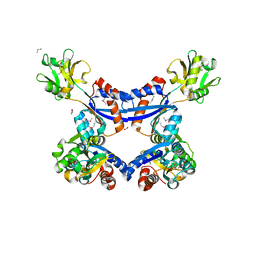 | | substrate binding domain of putative molybdenum ABC transporter from Clostridium difficile | | Descriptor: | 2-BUTANOL, ABC-type transport system, molybdenum-specific extracellular solute-binding protein, ... | | Authors: | Maltseva, N, Kim, Y, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-04-24 | | Release date: | 2013-05-08 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.4999 Å) | | Cite: | Substrate binding domain of putative molybdenum ABC transporter from Clostridium difficile 630
TO BE PUBLISHED
|
|
4KQ9
 
 | | Crystal structure of periplasmic ribose ABC transporter from Conexibacter woesei DSM 14684 | | Descriptor: | GLYCEROL, Ribose ABC transporter, substrate binding protein | | Authors: | Nocek, B, Chhor, G, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-14 | | Release date: | 2013-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of periplasmic ribose ABC transporter from Conexibacter woesei DSM 14684
To be Published
|
|
4KMR
 
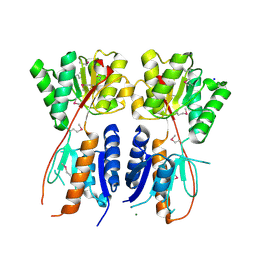 | | Structure of a putative transcriptional regulator of LacI family from Sanguibacter keddieii DSM 10542. | | Descriptor: | MAGNESIUM ION, SODIUM ION, Transcriptional regulator, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-08 | | Release date: | 2013-06-05 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a putative transcriptional regulator of LacI family from Sanguibacter keddieii DSM 10542.
To be Published
|
|
4MA0
 
 | | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with partially hydrolysed ATP | | Descriptor: | ADENOSINE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-15 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with partially hydrolysed ATP
To be Published
|
|
4M7O
 
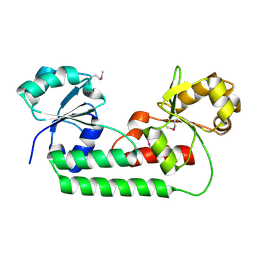 | |
4QPE
 
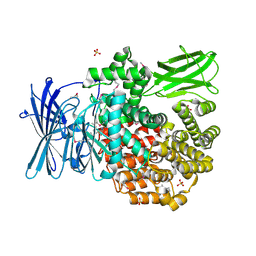 | | Crystal structure of Aminopeptidase N in complex with N-cyclohexyl-1,2-diaminoethylphosphonic acid | | Descriptor: | Aminopeptidase N, SULFATE ION, ZINC ION, ... | | Authors: | Nocek, B, Mulligan, R, Berlicki, L, Vassilious, S, Mucha, A, Joachimiak, A. | | Deposit date: | 2014-06-23 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Structure-guided, single-point modifications in the phosphinic dipeptide structure yield highly potent and selective inhibitors of neutral aminopeptidases.
J.Med.Chem., 57, 2014
|
|
4I9V
 
 | | The atomic structure of 5-Hydroxymethyl 2'-deoxycitidine base paired with 2'-deoxyguanosine in Dickerson Drew Dodecamer | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(5HC)P*GP*CP*G)-3'), MAGNESIUM ION, SPERMINE (FULLY PROTONATED FORM) | | Authors: | Nocek, B, Szulik, M.W, Joachimiak, A, Stone, M.P. | | Deposit date: | 2012-12-05 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Differential stabilities and sequence-dependent base pair opening dynamics of watson-crick base pairs with 5-hydroxymethylcytosine, 5-formylcytosine, or 5-carboxylcytosine.
Biochemistry, 54, 2015
|
|
4QME
 
 | | Crystal structure of Aminopeptidase N in complex with the phosphinic dipeptide analogue LL-(R,S)-hPheP[CH2]Phe | | Descriptor: | (2S)-3-[(S)-[(1R)-1-amino-3-phenylpropyl](hydroxy)phosphoryl]-2-benzylpropanoic acid, Aminopeptidase N, GLYCEROL, ... | | Authors: | Nocek, B, Vassilious, S, Mulligan, R, Berlicki, L, Mucha, A, Joachimiak, A. | | Deposit date: | 2014-06-16 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structure-guided, single-point modifications in the phosphinic dipeptide structure yield highly potent and selective inhibitors of neutral aminopeptidases.
J.Med.Chem., 57, 2014
|
|
4QN8
 
 | | The crystal structure of an effector protein VipE from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | Descriptor: | BETA-MERCAPTOETHANOL, VipE | | Authors: | Tan, K, Xu, X, Cui, H, Liu, S, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-06-17 | | Release date: | 2014-07-16 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | The crystal structure of an effector protein VipE from Legionella pneumophila subsp. pneumophila str. Philadelphia 1
To be Published
|
|
4MAM
 
 | | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with an ADP analog, AMP-CP | | Descriptor: | GLYCEROL, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, Phosphoribosylaminoimidazole carboxylase, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-16 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.474 Å) | | Cite: | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with an ADP analog, AMP-CP
To be Published
|
|
4KT7
 
 | | The crystal structure of 4-diphosphocytidyl-2C-methyl-D-erythritolsynthase from Anaerococcus prevotii DSM 20548 | | Descriptor: | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, CHLORIDE ION, SODIUM ION | | Authors: | Borek, D, Tan, K, Stols, L, Eschenfeidt, W.H, Otwinoski, Z, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-20 | | Release date: | 2013-06-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | The crystal structure of 4-diphosphocytidyl-2C-methyl-D-erythritolsynthase from Anaerococcus prevotii DSM 20548
To be Published
|
|
2P35
 
 | | Crystal structure of trans-aconitate methyltransferase from Agrobacterium tumefaciens | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, Trans-aconitate 2-methyltransferase | | Authors: | Chang, C, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-03-08 | | Release date: | 2007-04-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of trans-aconitate methyltransferase from Agrobacterium tumefaciens
To be Published
|
|
4ND9
 
 | | The putative substrate binding domain of ABC-type transporter from Agrobacterium tumefaciens in open conformation | | Descriptor: | ABC transporter, substrate binding protein (Proline/glycine/betaine) | | Authors: | Nicholls, R, Tkaczuk, K.L, Kagan, O, Chruszcz, M, Domagalski, M.J, Savchenko, A, Joachimiak, A, Murshudov, G, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-10-25 | | Release date: | 2013-12-11 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The putative substrate binding domain of ABC-type transporter from Agrobacterium tumefaciens in open conformation
To be Published
|
|
4M9U
 
 | | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | ACETATE ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-15 | | Release date: | 2013-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4
To be Published
|
|
4MDY
 
 | |
4LPQ
 
 | | Crystal structure of the L,D-transpeptidase (residues 123-326) from Xylanimonas cellulosilytica DSM 15894 | | Descriptor: | CHLORIDE ION, ErfK/YbiS/YcfS/YnhG family protein | | Authors: | Nocek, B, Bigelow, L, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-07-16 | | Release date: | 2013-11-13 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal structure of the L,D-transpeptidase (residues 123-326) from Xylanimonas cellulosilytica DSM 15894
To be Published
|
|
4MA5
 
 | | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with an ATP analog, AMP-PNP. | | Descriptor: | FORMIC ACID, GLYCEROL, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-15 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.809 Å) | | Cite: | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with an ATP analog, AMP-PNP.
To be Published
|
|
2OV9
 
 | | Crystal structure of protein RHA08564, thioesterase superfamily protein | | Descriptor: | Hypothetical protein, SULFATE ION | | Authors: | Chang, C, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-02-13 | | Release date: | 2007-03-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of protein RHA08564, thioesterase superfamily protein
To be Published
|
|
4LQB
 
 | | Crystal structure of uncharacterized protein Kfla3161 | | Descriptor: | CITRIC ACID, GLYCEROL, Uncharacterized protein | | Authors: | Chang, C, Chhor, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-07-17 | | Release date: | 2013-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of uncharacterized protein Kfla3161
To be Published
|
|
4M9D
 
 | | The Crystal structure of an adenylosuccinate synthetase from Bacillus anthracis str. Ames Ancestor in complex with AMP. | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Adenylosuccinate synthetase, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-14 | | Release date: | 2013-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | The Crystal structure of an adenylosuccinate synthetase from Bacillus anthracis str. Ames Ancestor in complex with AMP.
To be Published
|
|
4QUO
 
 | | Crystal structure of Aminopeptidase N in complex with the phosphinic dipeptide analogue LL-(R,S)-hPheP[CH2]Phe(3-CH2NH2) | | Descriptor: | (2S)-2-[3-(aminomethyl)benzyl]-3-[(R)-[(1R)-1-amino-3-phenylpropyl](hydroxy)phosphoryl]propanoic acid, Aminopeptidase N, GLYCEROL, ... | | Authors: | Nocek, B, Mulligan, R, Joachimiak, A, Vassiliou, S, Berlicki, L, Mucha, A. | | Deposit date: | 2014-07-11 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-guided, single-point modifications in the phosphinic dipeptide structure yield highly potent and selective inhibitors of neutral aminopeptidases.
J.Med.Chem., 57, 2014
|
|
