9AUW
 
 | | Crystal structure of A. baumannii GuaB dCBS with inhibitor GNE9979 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-[4-chloro-3-(dimethylamino)phenyl]-N~2~-[3-(hydroxymethyl)quinolin-6-yl]-L-alaninamide | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-03-01 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of GuaB inhibitors with efficacy against Acinetobacter baumannii infection.
Mbio, 15, 2024
|
|
9AUV
 
 | | Crystal structure of A. baumannii GuaB dCBS with inhibitor GNE9123 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-(6-chloropyridin-3-yl)-N~2~-(1,4-dihydro-2H-pyrano[3,4-c]quinolin-9-yl)-L-alaninamide | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-03-01 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of GuaB inhibitors with efficacy against Acinetobacter baumannii infection.
Mbio, 15, 2024
|
|
9AUX
 
 | | Crystal structure of A. baumannii GuaB dCBS with inhibitor GNE2011 | | Descriptor: | 9-{(1R)-1-[(5P)-5-(4-chloro-1H-imidazol-2-yl)pyridin-3-yl]ethoxy}-1,4-dihydro-2H-pyrano[3,4-c]quinoline, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-03-01 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Discovery of GuaB inhibitors with efficacy against Acinetobacter baumannii infection.
Mbio, 15, 2024
|
|
9AUZ
 
 | | Crystal structure of S. aureus GuaB dCBS with inhibitor GNE9979 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-[4-chloro-3-(dimethylamino)phenyl]-N~2~-[3-(hydroxymethyl)quinolin-6-yl]-L-alaninamide | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-03-01 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of GuaB inhibitors with efficacy against Acinetobacter baumannii infection.
Mbio, 15, 2024
|
|
9AV2
 
 | | Crystal structure of E. coli GuaB dCBS with inhibitor GNE9979 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-[4-chloro-3-(dimethylamino)phenyl]-N~2~-[3-(hydroxymethyl)quinolin-6-yl]-L-alaninamide | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-03-01 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of GuaB inhibitors with efficacy against Acinetobacter baumannii infection.
Mbio, 15, 2024
|
|
9AUY
 
 | | Crystal structure of S. aureus GuaB dCBS with inhibitor GNE9123 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-(6-chloropyridin-3-yl)-N~2~-(1,4-dihydro-2H-pyrano[3,4-c]quinolin-9-yl)-L-alaninamide | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-03-01 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of GuaB inhibitors with efficacy against Acinetobacter baumannii infection.
Mbio, 15, 2024
|
|
9AV1
 
 | | Crystal structure of E. coli GuaB dCBS with inhibitor GNE9123 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-(6-chloropyridin-3-yl)-N~2~-(1,4-dihydro-2H-pyrano[3,4-c]quinolin-9-yl)-L-alaninamide | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-03-01 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of GuaB inhibitors with efficacy against Acinetobacter baumannii infection.
Mbio, 15, 2024
|
|
9AV3
 
 | | Crystal structure of E. coli GuaB dCBS with inhibitor GNE2011 | | Descriptor: | 9-{(1R)-1-[(5P)-5-(4-chloro-1H-imidazol-2-yl)pyridin-3-yl]ethoxy}-1,4-dihydro-2H-pyrano[3,4-c]quinoline, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-03-01 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Discovery of GuaB inhibitors with efficacy against Acinetobacter baumannii infection.
Mbio, 15, 2024
|
|
5XKF
 
 | | Crystal structure of T2R-TTL-MPC6827 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Y, Yang, J, Wang, T, Chen, L. | | Deposit date: | 2017-05-07 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of a powerful and reversible microtubule-inhibitor with efficacy against multidrug-resistant tumors
To Be Published
|
|
6KN5
 
 | |
8WQO
 
 | | Crystal structure of BRD4-BD1 bound with DI106 | | Descriptor: | (3~{R})-6-[[4-(3,5-dimethyl-1~{H}-pyrazol-4-yl)pyrimidin-2-yl]amino]-1,3-dimethyl-4-propan-2-yl-3~{H}-quinoxalin-2-one, Isoform C of Bromodomain-containing protein 4 | | Authors: | Cao, D, Xiong, B. | | Deposit date: | 2023-10-12 | | Release date: | 2024-07-24 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Structure-Based Rational Design and Evaluation of BET-Aurora Kinase Dual-Inhibitors for Treatment of Cancers.
J.Med.Chem., 68, 2025
|
|
5XKE
 
 | | Crystal structure of T2R-TTL-Demecolcine complex | | Descriptor: | (7S)-1,2,3,10-tetramethoxy-7-(methylamino)-6,7-dihydro-5H-benzo[a]heptalen-9-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Yang, J, Wang, T, Chen, L. | | Deposit date: | 2017-05-07 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of a powerful and reversible microtubule-inhibitor with efficacy against multidrug-resistant tumors
To Be Published
|
|
3G9I
 
 | | GR DNA Binding domain: Pal complex-35 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*AP*GP*AP*AP*CP*AP*TP*TP*TP*TP*GP*TP*TP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*AP*AP*CP*AP*AP*AP*AP*TP*GP*TP*TP*CP*T)-3'), ... | | Authors: | Pufall, M.A, Yamamoto, K.R, Meijsing, S.H. | | Deposit date: | 2009-02-13 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | DNA binding site sequence directs glucocorticoid receptor structure and activity.
Science, 324, 2009
|
|
3HC5
 
 | | FXR with SRC1 and GSK826 | | Descriptor: | 3-(6-{[3-(2,6-dichlorophenyl)-5-(1-methylethyl)isoxazol-4-yl]methoxy}-1-benzothiophen-2-yl)benzoic acid, Bile acid receptor, Nuclear receptor coactivator 1, ... | | Authors: | Williams, S.P, Madauss, K.P. | | Deposit date: | 2009-05-05 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | FXR agonist activity of conformationally constrained analogs of GW 4064.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2GTG
 
 | |
2DOB
 
 | | Crystal Structure of Human Saposin A | | Descriptor: | CALCIUM ION, Proactivator polypeptide | | Authors: | Prive, G.G, Ahn, V.E. | | Deposit date: | 2006-04-28 | | Release date: | 2006-07-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of saposins A and C.
Protein Sci., 15, 2006
|
|
2V6Q
 
 | |
2EFL
 
 | | Crystal structure of the EFC domain of formin-binding protein 17 | | Descriptor: | Formin-binding protein 1 | | Authors: | Shimada, A, Niwa, H, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-23 | | Release date: | 2007-05-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Curved EFC/F-BAR-Domain Dimers Are Joined End to End into a Filament for Membrane Invagination in Endocytosis
Cell(Cambridge,Mass.), 129, 2007
|
|
7VSQ
 
 | |
7VSP
 
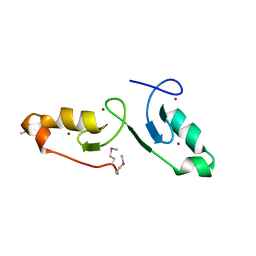 | | Crystal strcuture of the tandem B-Box domains of COL2 | | Descriptor: | ZINC ION, Zinc finger protein CONSTANS-LIKE 2 | | Authors: | Lv, X, Liu, R, Du, J. | | Deposit date: | 2021-10-27 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis of CONSTANS oligomerization in FLOWERING LOCUS T activation.
J Integr Plant Biol, 64, 2022
|
|
7VLA
 
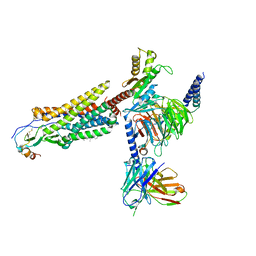 | | Cryo-EM structure of the CCL15(27-92) bound CCR1-Gi complex | | Descriptor: | C-C chemokine receptor type 1, CCL15(27-92), CHOLESTEROL, ... | | Authors: | Shao, Z, Shen, Q, Mao, C, Yao, B, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Ma, H, Chen, Z, Xu, H.E, Ying, S, Zhang, Y, Shen, H. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Identification and mechanism of G protein-biased ligands for chemokine receptor CCR1.
Nat.Chem.Biol., 18, 2022
|
|
7VL8
 
 | | Cryo-EM structure of the Apo CCR1-Gi complex | | Descriptor: | C-C chemokine receptor type 1, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shao, Z, Shen, Q, Mao, C, Yao, B, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Ma, H, Chen, Z, Xu, H.E, Ying, S, Zhang, Y, Shen, H. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-23 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Identification and mechanism of G protein-biased ligands for chemokine receptor CCR1.
Nat.Chem.Biol., 18, 2022
|
|
7VL9
 
 | | Cryo-EM structure of the CCL15(26-92) bound CCR1-Gi complex | | Descriptor: | C-C chemokine receptor type 1, CCL15(26-92), CHOLESTEROL, ... | | Authors: | Shao, Z, Shen, Q, Mao, C, Yao, B, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Ma, H, Chen, Z, Xu, H.E, Ying, S, Zhang, Y, Shen, H. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-23 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Identification and mechanism of G protein-biased ligands for chemokine receptor CCR1.
Nat.Chem.Biol., 18, 2022
|
|
6NF0
 
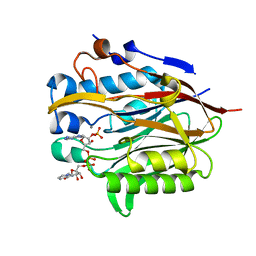 | | Nocturnin with bound NADPH substrate | | Descriptor: | CALCIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Nocturnin | | Authors: | Estrella, M.A, Du, J, Korennykh, A. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The metabolites NADP+and NADPH are the targets of the circadian protein Nocturnin (Curled).
Nat Commun, 10, 2019
|
|
3M5B
 
 | |
