4ZY4
 
 | |
2FX6
 
 | | bovine trypsin complexed with 2-aminobenzamidazole | | Descriptor: | 2H-BENZOIMIDAZOL-2-YLAMINE, CALCIUM ION, Trypsin | | Authors: | Katz, B.A. | | Deposit date: | 2006-02-03 | | Release date: | 2006-02-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structure-guided design of Peptide-based tryptase inhibitors.
Biochemistry, 45, 2006
|
|
2FWW
 
 | |
6V3N
 
 | | Crystal structure of CDYL2 in complex with H3K27me3 | | Descriptor: | ACE-GLN-LEU-ALA-THR-LYS-ALA-ALA-ARG-M3L-SER-ALA-PRO-ALA-THR-TYR-NH2, Chromodomain Y-like protein 2, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
2FPZ
 
 | | Human tryptase with 2-amino benzimidazole | | Descriptor: | 2H-BENZOIMIDAZOL-2-YLAMINE, Tryptase beta-2 | | Authors: | Somoza, J.R. | | Deposit date: | 2006-01-17 | | Release date: | 2006-03-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided design of Peptide-based tryptase inhibitors.
Biochemistry, 45, 2006
|
|
2FS8
 
 | | Human beta-tryptase II with inhibitor CRA-29382 | | Descriptor: | ALLYL {(1S)-1-[(5-{4-[(2,3-DIHYDRO-1H-INDEN-2-YLAMINO)CARBONYL]BENZYL}-1,2,4-OXADIAZOL-3-YL)CARBONYL]-3-PYRROLIDIN-3-YLPROPYL}CARBAMATE, Tryptase beta-2 | | Authors: | Somoza, J.R. | | Deposit date: | 2006-01-21 | | Release date: | 2006-03-21 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-guided design of Peptide-based tryptase inhibitors.
Biochemistry, 45, 2006
|
|
4ZY5
 
 | |
6O3N
 
 | |
4PNE
 
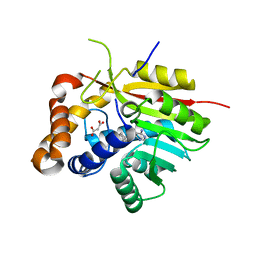 | | Crystal Structure of the [4+2]-Cyclase SpnF | | Descriptor: | MALONATE ION, Methyltransferase-like protein, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Fage, C.D, Isiorho, E.A, Liu, Y.-N, Liu, H.-W, Keatinge-Clay, A.T. | | Deposit date: | 2014-05-23 | | Release date: | 2015-02-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of SpnF, a standalone enzyme that catalyzes [4 + 2] cycloaddition.
Nat.Chem.Biol., 11, 2015
|
|
5TCZ
 
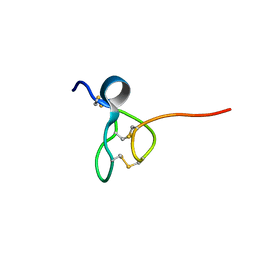 | |
3N9M
 
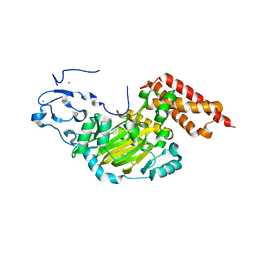 | | ceKDM7A from C.elegans, alone | | Descriptor: | FE (II) ION, Putative uncharacterized protein, ZINC ION | | Authors: | Yang, Y, Hu, L, Wang, P, Hou, H, Chen, C.D, Xu, Y. | | Deposit date: | 2010-05-31 | | Release date: | 2010-06-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Structural insights into a dual-specificity histone demethylase ceKDM7A from Caenorhabditis elegans
Cell Res., 20, 2010
|
|
2FS9
 
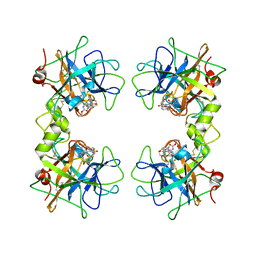 | | Human beta tryptase II with inhibitor CRA-28427 | | Descriptor: | ETHYL {(1S)-5-AMINO-1-[(5-{4-[(2,3-DIHYDRO-1H-INDEN-2-YLAMINO)CARBONYL]BENZYL}-1,2,4-OXADIAZOL-3-YL)CARBONYL]PENTYL}CARBAMATE, Tryptase beta-2 | | Authors: | Somoza, J.R. | | Deposit date: | 2006-01-21 | | Release date: | 2006-03-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided design of Peptide-based tryptase inhibitors.
Biochemistry, 45, 2006
|
|
2FXR
 
 | | human beta tryptase II complexed with activated ketone inhibitor CRA-29382 | | Descriptor: | ALLYL {(1S)-1-[(5-{4-[(2,3-DIHYDRO-1H-INDEN-2-YLAMINO)CARBONYL]BENZYL}-1,2,4-OXADIAZOL-3-YL)CARBONYL]-3-PYRROLIDIN-3-YLPROPYL}CARBAMATE, Tryptase beta-2 | | Authors: | Katz, B.A. | | Deposit date: | 2006-02-06 | | Release date: | 2006-02-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-guided design of Peptide-based tryptase inhibitors.
Biochemistry, 45, 2006
|
|
6BAG
 
 | | Lactate Dehydrogenase in complex with inhibitor (R)-5-((2-chlorophenyl)thio)-6'-((4-fluorophenyl)amino)-4-hydroxy-2-(thiophen-3-yl)-2,3-dihydro-[2,2'-bipyridin]-6(1H)-one | | Descriptor: | (2R)-5-[(2-chlorophenyl)sulfanyl]-6'-[(4-fluorophenyl)amino]-4-hydroxy-2-(thiophen-3-yl)-2,3-dihydro[2,2'-bipyridin]-6(1H)-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, L-lactate dehydrogenase A chain, ... | | Authors: | Ultsch, M, Eigenbrot, C. | | Deposit date: | 2017-10-12 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-guided optimization and in vivo activities of hydroxylactone and hydroxylactam Inhibitors of Human Lactate Dehydrogenase
To Be Published
|
|
6BB0
 
 | |
7EXA
 
 | | Structure of mumps virus nucleoprotein without C-arm | | Descriptor: | Nucleoprotein, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') | | Authors: | Shen, Q, Shan, H, Zhang, N, Qin, Y. | | Deposit date: | 2021-05-26 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural plasticity of mumps virus nucleocapsids with cryo-EM structures.
Commun Biol, 4, 2021
|
|
6BAZ
 
 | | Lactate Dehydrogenase in complex with inhibitor (S)-5-((2-chlorophenyl)thio)-6'-((4-fluorophenyl)amino)-4-hydroxy-2-(thiophen-3-yl)-2,3-dihydro-[2,2'-bipyridin]-6(1H)-one | | Descriptor: | (3S,6S)-3-[(2-chlorophenyl)sulfanyl]-6-{6-[(4-fluorophenyl)amino]pyridin-2-yl}-6-(thiophen-3-yl)piperidine-2,4-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, L-lactate dehydrogenase A chain, ... | | Authors: | Ultsch, M, Eigenbrot, C. | | Deposit date: | 2017-10-16 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-guided optimization and in vivo activities of hydroxylactone and hydroxylactam Inhibitors of Human Lactate Dehydrogenase
To Be Published
|
|
6BAD
 
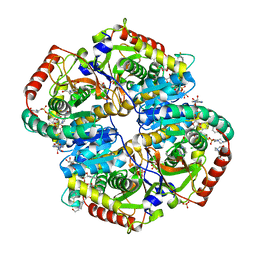 | |
4R89
 
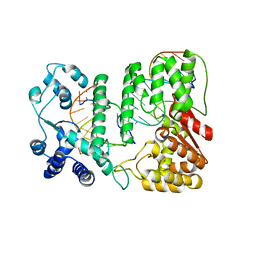 | | Crystal structure of paFAN1 - 5' flap DNA complex with Manganase | | Descriptor: | DNA (5'-D(P*AP*CP*CP*AP*GP*AP*CP*AP*CP*AP*CP*AP*TP*TP*C)-3'), DNA (5'-D(P*GP*AP*AP*TP*GP*TP*GP*TP*GP*TP*CP*TP*CP*AP*AP*TP*CP*CP*CP*AP*AP*C)-3'), DNA (5'-D(P*GP*TP*TP*GP*GP*GP*AP*TP*TP*G)-3'), ... | | Authors: | Cho, Y, Gwon, G.H, Kim, Y.R. | | Deposit date: | 2014-08-30 | | Release date: | 2014-10-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (4.002 Å) | | Cite: | Crystal structure of a Fanconi anemia-associated nuclease homolog bound to 5' flap DNA: basis of interstrand cross-link repair by FAN1
Genes Dev., 28, 2014
|
|
8WB6
 
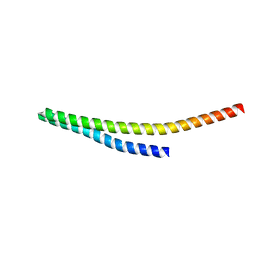 | |
8WB7
 
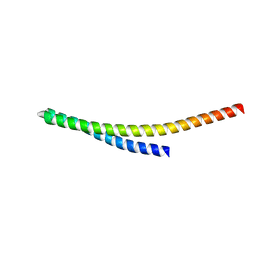 | |
4R8A
 
 | | Crystal structure of paFAN1 - 5' flap DNA complex | | Descriptor: | DNA (5'-D(P*AP*CP*CP*AP*GP*AP*CP*AP*CP*AP*CP*AP*TP*TP*C)-3'), DNA (5'-D(P*GP*AP*AP*TP*GP*TP*GP*TP*GP*TP*CP*TP*CP*AP*AP*TP*CP*CP*CP*AP*A)-3'), DNA (5'-D(P*GP*TP*TP*GP*GP*GP*AP*TP*TP*G)-3'), ... | | Authors: | Cho, Y, Gwon, G.H, Kim, Y.R. | | Deposit date: | 2014-08-30 | | Release date: | 2014-10-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a Fanconi anemia-associated nuclease homolog bound to 5' flap DNA: basis of interstrand cross-link repair by FAN1
Genes Dev., 28, 2014
|
|
6IXM
 
 | | Crystal structure of the ketone reductase ChKRED20 from the genome of Chryseobacterium sp. CA49 complexed with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Short-chain dehydrogenase reductase | | Authors: | Zhao, F.J, Jin, Y, Liu, Z.C, Wang, G.G, Wu, Z.L. | | Deposit date: | 2018-12-11 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structure-guided engineering of ChKRED20 from Chryseobacterium sp. CA49 for asymmetric reduction of aryl ketoesters.
Enzyme Microb. Technol., 125, 2019
|
|
7TXI
 
 | | Cryo-EM of A. pernix flagellum | | Descriptor: | Probable flagellin 1 | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Baquero, D.P, Krupovic, M, Egelman, E.H. | | Deposit date: | 2022-02-09 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The evolution of archaeal flagellar filaments.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3HW3
 
 | | The crystal structure of avian influenza virus PA_N in complex with UMP | | Descriptor: | MAGNESIUM ION, Polymerase acidic protein, URIDINE-5'-MONOPHOSPHATE | | Authors: | Zhao, C, Lou, Z, Guo, Y, Ma, M, Chen, Y, Rao, Z. | | Deposit date: | 2009-06-17 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nucleoside monophosphate complex structures of the endonuclease domain from the influenza virus polymerase PA subunit reveal the substrate binding site inside the catalytic center
J.Virol., 83, 2009
|
|
