6IFR
 
 | | Type III-A Csm complex, Cryo-EM structure of Csm-NTR, ATP bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Type III-A CRISPR-associated RAMP protein Csm3, ... | | Authors: | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | Deposit date: | 2018-09-21 | | Release date: | 2018-12-12 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
6IFK
 
 | | Cryo-EM structure of type III-A Csm-CTR1 complex, AMPPNP bound | | Descriptor: | CTR1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | Deposit date: | 2018-09-20 | | Release date: | 2018-12-12 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
6IFL
 
 | | Cryo-EM structure of type III-A Csm-NTR complex | | Descriptor: | NTR, Type III-A CRISPR-associated RAMP protein Csm3, Type III-A CRISPR-associated RAMP protein Csm4, ... | | Authors: | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | Deposit date: | 2018-09-20 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
6IFY
 
 | | Type III-A Csm complex, Cryo-EM structure of Csm-CTR1 | | Descriptor: | CTR1, Type III-A CRISPR-associated RAMP protein Csm3, Type III-A CRISPR-associated RAMP protein Csm4, ... | | Authors: | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | Deposit date: | 2018-09-21 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
6HTS
 
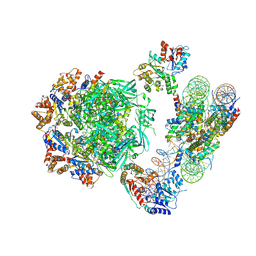 | | Cryo-EM structure of the human INO80 complex bound to nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling ATPase INO80, ... | | Authors: | Ayala, R, Willhoft, O, Aramayo, R.J, Wilkinson, M, McCormack, E.A, Ocloo, L, Wigley, D.B, Zhang, X. | | Deposit date: | 2018-10-04 | | Release date: | 2018-11-07 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure and regulation of the human INO80-nucleosome complex.
Nature, 556, 2018
|
|
6IG0
 
 | | Type III-A Csm complex, Cryo-EM structure of Csm-CTR1, ATP bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CTR1, MAGNESIUM ION, ... | | Authors: | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | Deposit date: | 2018-09-21 | | Release date: | 2018-12-12 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
6I52
 
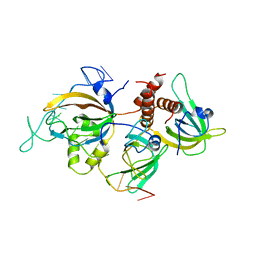 | | Yeast RPA bound to ssDNA | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Replication factor A protein 1, Replication factor A protein 2, ... | | Authors: | Yates, L.A, Aramayo, R.J, Zhang, X. | | Deposit date: | 2018-11-12 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | A structural and dynamic model for the assembly of Replication Protein A on single-stranded DNA.
Nat Commun, 9, 2018
|
|
6KA4
 
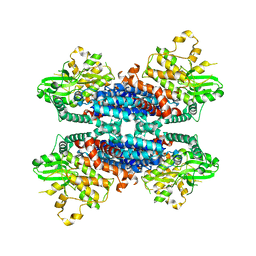 | | Cryo-EM structure of the AtMLKL3 tetramer | | Descriptor: | F22L4.1 protein | | Authors: | Lisa, M, Huang, M, Zhang, X, Ryohei, T.N, Leila, B.K, Isabel, M.L.S, Florence, J, Viera, K, Dmitry, L, Jane, E.P, James, M.M, Kay, H, Paul, S.L, Chai, J, Takaki, M. | | Deposit date: | 2019-06-20 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the AtMLKL3 tetramer
To Be Published
|
|
6IFU
 
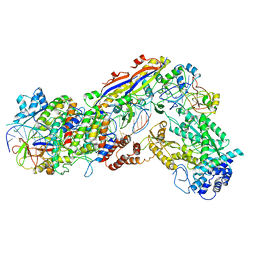 | | Cryo-EM structure of type III-A Csm-CTR2-dsDNA complex | | Descriptor: | CTR2, Type III-A CRISPR-associated RAMP protein Csm3, Type III-A CRISPR-associated RAMP protein Csm4, ... | | Authors: | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | Deposit date: | 2018-09-21 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
6HT4
 
 | |
5XGR
 
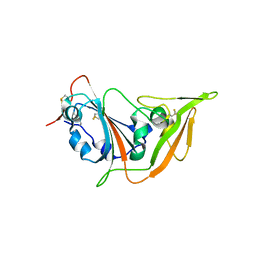 | | Structure of the S1 subunit C-terminal domain from bat-derived coronavirus HKU5 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1 | | Authors: | Xue, H, Qi, J, Song, H, Qihui, W, Shi, Y, Gao, G.F. | | Deposit date: | 2017-04-16 | | Release date: | 2017-05-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the S1 subunit C-terminal domain from bat-derived coronavirus HKU5 spike protein
Virology, 507, 2017
|
|
9J0J
 
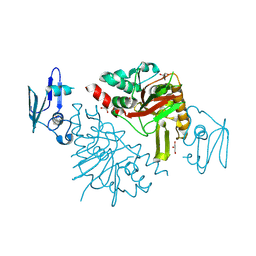 | | The crystal structure of Fe/2OG-dependent oxygenase DfmD | | Descriptor: | FE (III) ION, Fe/2OG-dependent oxygenase DfmD, GLYCEROL, ... | | Authors: | Wang, L, Chen, J, Zhou, J. | | Deposit date: | 2024-08-02 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | An Alternative Mechanism for C-C Desaturation Underscores a Dual-Controlled Mechanism for the Fate of Radical Intermediate in Iron(II)- and 2-(Oxo)glutarate-Dependent Oxygenase DfmD.
J.Am.Chem.Soc., 147, 2025
|
|
5XS2
 
 | |
5XQX
 
 | |
9M7V
 
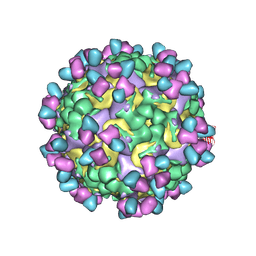 | | Cryo-EM structure of enterovirus A71 mature virion in complex with Fab CT11F9 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Jiang, Y, Zhu, R, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2025-03-11 | | Release date: | 2025-04-23 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Development of In Vitro Potency Methods to Replace In Vivo Tests for Enterovirus 71 Inactivated Vaccine (Human Diploid Cell-Based/Vero Cell-Based).
Vaccines (Basel), 13, 2025
|
|
3J35
 
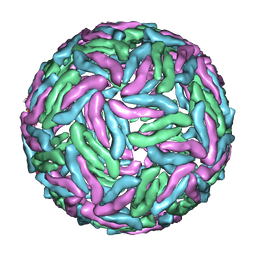 | | Cryo-EM reconstruction of Dengue virus at 37 C | | Descriptor: | envelope protein | | Authors: | Zhang, X.Z, Sheng, J, Plevka, P, Kuhn, R.J, Diamond, M.S, Rossmann, M.G. | | Deposit date: | 2013-02-24 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | Dengue structure differs at the temperatures of its human and mosquito hosts.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1L71
 
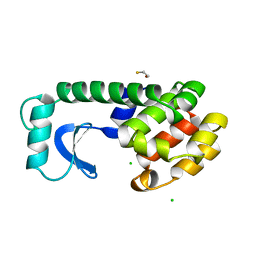 | |
1L69
 
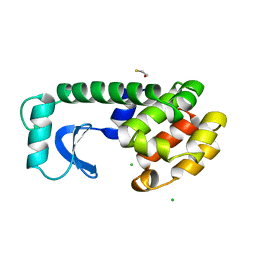 | |
5JRH
 
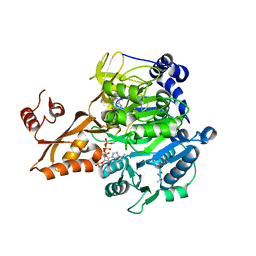 | | Crystal structure of Salmonella enterica acetyl-CoA synthetase (Acs) in complex with cAMP and Coenzyme A | | Descriptor: | (R,R)-2,3-BUTANEDIOL, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Acetyl-coenzyme A synthetase, ... | | Authors: | Shen, L, Zhang, Y. | | Deposit date: | 2016-05-06 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.644 Å) | | Cite: | Cyclic AMP Inhibits the Activity and Promotes the Acetylation of Acetyl-CoA Synthetase through Competitive Binding to the ATP/AMP Pocket.
J. Biol. Chem., 292, 2017
|
|
6FPF
 
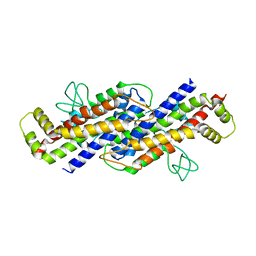 | |
6FPG
 
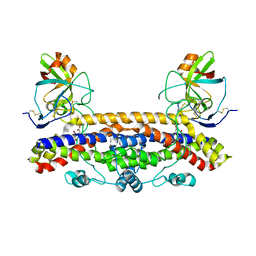 | | Structure of the Ustilago maydis chorismate mutase 1 in complex with a Zea mays kiwellin | | Descriptor: | CITRIC ACID, Chromosome 16, whole genome shotgun sequence, ... | | Authors: | Altegoer, F, Steinchen, W, Bange, G. | | Deposit date: | 2018-02-09 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A kiwellin disarms the metabolic activity of a secreted fungal virulence factor.
Nature, 565, 2019
|
|
6H3P
 
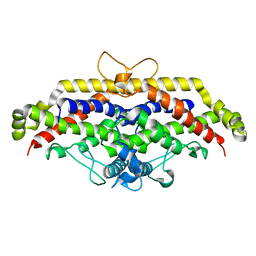 | |
8XFD
 
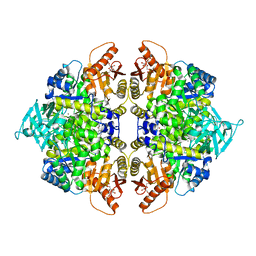 | | Crystal structure of pyruvate kinase tetramer in complex with allosteric activator, Mitapivat (MTPV, AG-348) | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Isoform L-type of Pyruvate kinase PKLR, MAGNESIUM ION, ... | | Authors: | Sun, R, Achour, A. | | Deposit date: | 2023-12-13 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High Resolution Crystal Structure of the Pyruvate Kinase Tetramer in Complex with the Allosteric Activator Mitapivat/AG-348
Crystals, 2024
|
|
6HJW
 
 | |
8WCT
 
 | |
