4UTB
 
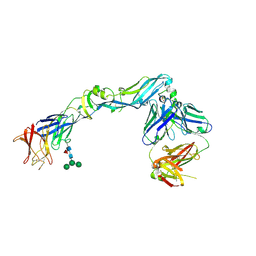 | | Crystal structure of dengue 2 virus envelope glycoprotein in complex with the Fab fragment of the broadly neutralizing human antibody EDE2 A11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 A11, ... | | Authors: | Rouvinski, A, Guardado-Calvo, P, Barba-Spaeth, G, Duquerroy, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2014-07-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Recognition Determinants of Broadly Neutralizing Human Antibodies Against Dengue Viruses.
Nature, 520, 2015
|
|
8SSU
 
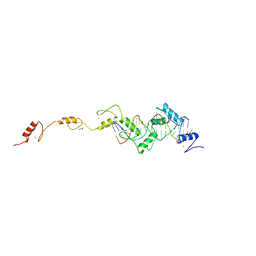 | | ZnFs 3-11 of CCCTC-binding factor (CTCF) Complexed with 19mer DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (19-MER) Strand I, DNA (19-MER) Strand II, ... | | Authors: | Horton, J.R, Yang, J, Cheng, X. | | Deposit date: | 2023-05-08 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structures of CTCF-DNA complexes including all 11 zinc fingers.
Nucleic Acids Res., 51, 2023
|
|
9BKN
 
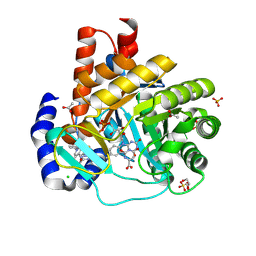 | | DHODH in complex with Ligand 16 | | Descriptor: | (2P,6P)-6-[4-ethyl-3-(hydroxymethyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl]-7-fluoro-2-(2-methylphenyl)-4-(propan-2-yl)isoquinolin-1(2H)-one, ACETATE ION, CHLORIDE ION, ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2024-04-29 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of JNJ-74856665: A Novel Isoquinolinone DHODH Inhibitor for the Treatment of AML.
J.Med.Chem., 2024
|
|
9BKO
 
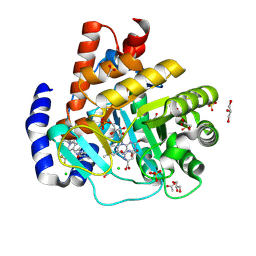 | | DHODH in complex with Ligand 26 | | Descriptor: | (2P,6P)-6-[4-ethyl-3-(hydroxymethyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl]-7-fluoro-2-(2-methylphenyl)-4-[(2R)-1,1,1-trifluoropropan-2-yl]isoquinolin-1(2H)-one, ACETATE ION, CHLORIDE ION, ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2024-04-29 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Discovery of JNJ-74856665: A Novel Isoquinolinone DHODH Inhibitor for the Treatment of AML.
J.Med.Chem., 2024
|
|
9BKM
 
 | | DHODH in complex with Ligand 10 | | Descriptor: | (2M,6P)-2-(2-chloro-6-fluorophenyl)-6-[4-ethyl-3-(hydroxymethyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl]-7-fluoro-4-(propan-2-yl)isoquinolin-1(2H)-one, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2024-04-29 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of JNJ-74856665: A Novel Isoquinolinone DHODH Inhibitor for the Treatment of AML.
J.Med.Chem., 2024
|
|
6DQB
 
 | | LINKED KDM5A JMJ DOMAIN FORMING COVALENT BOND TO INHIBITOR N71 i.e. 2-((3-(4-(dimethylamino)but-2-enamido)phenyl)(2-(piperidin-1-yl)ethoxy)methyl)thieno[3,2-b]pyridine-7-carboxylic acid | | Descriptor: | 2-{(R)-(3-{[(2E)-4-(dimethylamino)but-2-enoyl]amino}phenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, 2-{(R)-(3-{[4-(dimethylamino)butanoyl]amino}phenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, 2-{(S)-(3-{[4-(dimethylamino)butanoyl]amino}phenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
6DQ8
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR N49 i.e. 2-((2-chlorophenyl)(2-(1-methylpyrrolidin-2-yl)ethoxy)methyl)thieno[3,2-b]pyridine-7-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-[(R)-(2-chlorophenyl){2-[(2S)-1-methylpyrrolidin-2-yl]ethoxy}methyl]thieno[3,2-b]pyridine-7-carboxylic acid, 2-[(S)-(2-chlorophenyl){2-[(2S)-1-methylpyrrolidin-2-yl]ethoxy}methyl]thieno[3,2-b]pyridine-7-carboxylic acid, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
6DQ6
 
 | |
6DQF
 
 | |
6DQA
 
 | | Linked KDM5A JMJ Domain Bound to Inhibitor N70 i.e.[2-((3-aminophenyl)(2-(piperidin-1-yl)ethoxy)methyl)thieno[3,2-b]pyridine-7-carboxylic acid] | | Descriptor: | 1,2-ETHANEDIOL, 2-{(R)-(3-aminophenyl)[2-(piperidin-1-yl)ethoxy]methyl}thieno[3,2-b]pyridine-7-carboxylic acid, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2018-06-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.888 Å) | | Cite: | Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A.
J. Med. Chem., 61, 2018
|
|
6DQC
 
 | |
6DQ7
 
 | |
8SEI
 
 | | SF Tau from Down Syndrome | | Descriptor: | Microtubule-associated protein tau | | Authors: | Hoq, M.R, Bharath, S.R, Jiang, W, Vago, F.S, Bharath, S.R. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-03 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of amyloid-beta and tau filaments in Down syndrome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8SEJ
 
 | |
8SEH
 
 | | PHF Tau from Down Syndrome | | Descriptor: | Microtubule-associated protein tau | | Authors: | Hoq, M.R, Bharath, S.R, Jiang, W, Vago, F.S. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-03 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of amyloid-beta and tau filaments in Down syndrome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8SEK
 
 | |
8SEL
 
 | |
7XOE
 
 | | Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Prefusion state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,peptide | | Authors: | Wu, Z, Yu, Z, Tan, S, Lu, J, Lu, G, Lin, J. | | Deposit date: | 2022-05-01 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Preclinical evaluation of RQ3013, a broad-spectrum mRNA vaccine against SARS-CoV-2 variants.
Sci Bull (Beijing), 68, 2023
|
|
7XOG
 
 | | Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Postfusion state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,peptide, ... | | Authors: | Wu, Z, Yu, Z, Tan, S, Lu, J, Lu, G, Lin, J. | | Deposit date: | 2022-05-01 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Preclinical evaluation of RQ3013, a broad-spectrum mRNA vaccine against SARS-CoV-2 variants.
Sci Bull (Beijing), 68, 2023
|
|
6KIF
 
 | | Structure of cyanobacterial photosystem I-IsiA-flavodoxin supercomplex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Cao, P, Cao, D.F, Si, L, Su, X.D, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for energy and electron transfer of the photosystem I-IsiA-flavodoxin supercomplex.
Nat.Plants, 6, 2020
|
|
3UI7
 
 | | Discovery of orally active pyrazoloquinoline as a potent PDE10 inhibitor for the management of schizophrenia | | Descriptor: | 6-methoxy-3,8-dimethyl-4-(morpholin-4-ylmethyl)-1H-pyrazolo[3,4-b]quinoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Yang, S, Smotryski, J, Mcelroy, W, Ho, G, Tulshian, D, Greenlee, W.J, Hodgson, R, Xiao, L, Hruza, A. | | Deposit date: | 2011-11-04 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery of orally active pyrazoloquinolines as potent PDE10 inhibitors for the management of schizophrenia.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
7XPO
 
 | | Crystal Structure of UDP-Glc/GlcNAc 4-Epimerase with NAD/UDP-Glc | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase, ... | | Authors: | Chen, Y.H, Wang, X.C, Zhang, C.R. | | Deposit date: | 2022-05-05 | | Release date: | 2023-05-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A maize epimerase modulates cell wall synthesis and glycosylation during stomatal morphogenesis.
Nat Commun, 14, 2023
|
|
7RN0
 
 | |
7RN1
 
 | |
7XED
 
 | | Crystal Structure of OsCIE1-Ubox and OsUBC8 complex | | Descriptor: | U-box domain-containing protein 12, UBC core domain-containing protein | | Authors: | Zhang, Y, Yu, C.Z. | | Deposit date: | 2022-03-30 | | Release date: | 2023-10-04 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Release of a ubiquitin brake activates OsCERK1-triggered immunity in rice.
Nature, 629, 2024
|
|
