6H8S
 
 | |
5BUG
 
 | | Crystal structure of human phosphatase PTEN oxidized by H2O2 | | Descriptor: | L(+)-TARTARIC ACID, Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN | | Authors: | Lee, C.-U, Bier, D, Hennig, S, Grossmann, T.N. | | Deposit date: | 2015-06-03 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Redox Modulation of PTEN Phosphatase Activity by Hydrogen Peroxide and Bisperoxidovanadium Complexes.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5B5Y
 
 | | Crystal structure of PtLCIB4, a homolog of the limiting CO2-inducible protein LCIB | | Descriptor: | ACETATE ION, PtLCIB4, ZINC ION | | Authors: | Jin, S, Sun, J, Wunder, T, Tang, D, Mueller-Cajar, O.M, Gao, Y. | | Deposit date: | 2016-05-24 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into the LCIB protein family reveals a new group of beta-carbonic anhydrases
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
1L8N
 
 | | The 1.5A crystal structure of alpha-D-glucuronidase from Bacillus stearothermophilus T-1, complexed with 4-O-methyl-glucuronic acid and xylotriose | | Descriptor: | 4-O-methyl-beta-D-glucopyranuronic acid, ALPHA-D-GLUCURONIDASE, GLYCEROL, ... | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2002-03-21 | | Release date: | 2003-03-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of Geobacillus stearothermophilus {alpha}-Glucuronidase Complexed with Its Substrate and Products: MECHANISTIC IMPLICATIONS.
J.Biol.Chem., 279, 2004
|
|
6HEW
 
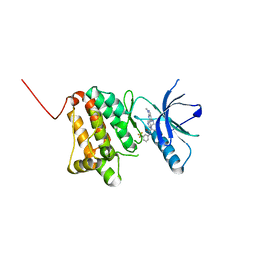 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with the NVP-BHG712 derivative AT069 | | Descriptor: | 4-methyl-3-[(1-methyl-6-pyrimidin-5-yl-pyrazolo[3,4-d]pyrimidin-4-yl)amino]-~{N}-[3-(trifluoromethyl)phenyl]benzamide, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Troester, A, Witt, K, Linhard, V.L, Gande, S.L, Saxena, K, Schwalbe, H. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.268 Å) | | Cite: | Effects of NVP-BHG712 chemical modifications on EPHA2 binding and affinity
To Be Published
|
|
6HF1
 
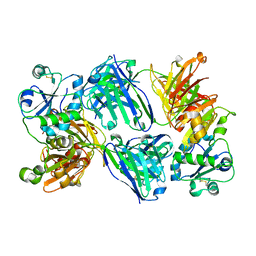 | |
5BPN
 
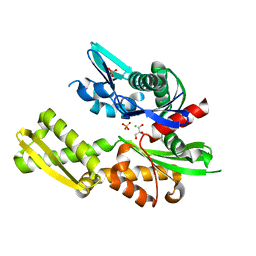 | |
6HG8
 
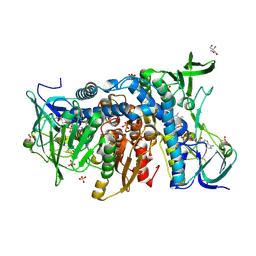 | | Crystal structure of the R460G disease-causing mutant of the human dihydrolipoamide dehydrogenase. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dihydrolipoyl dehydrogenase, mitochondrial, ... | | Authors: | Ambrus, A, Szabo, E, Weichsel, A, Bui, D, Wilk, P, Torocsik, B, Weiss, M.S, Montfort, W.R, Jordan, F, Adam-Vizi, V. | | Deposit date: | 2018-08-22 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of the R460G disease-causing mutant of the human dihydrolipoamide dehydrogenase.
To Be Published
|
|
5BRB
 
 | | Crystal structure of Q64E mutant of Triosephosphate isomerase from Plasmodium falciparum | | Descriptor: | SODIUM ION, Triosephosphate isomerase | | Authors: | Bandyopadhyay, D, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2015-05-30 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Probing the role of highly conserved residues in triosephosphate isomerase - analysis of site specific mutants at positions 64 and 75 in the Plasmodial enzyme
Febs J., 282, 2015
|
|
6HHF
 
 | | Crystal Structure of AKT1 in Complex with Covalent-Allosteric AKT Inhibitor Borussertib | | Descriptor: | Borussertib, RAC-alpha serine/threonine-protein kinase | | Authors: | Landel, I, Weisner, J, Mueller, M.P, Scheinpflug, R, Rauh, D. | | Deposit date: | 2018-08-28 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Preclinical Efficacy of Covalent-Allosteric AKT Inhibitor Borussertib in Combination with Trametinib inKRAS-Mutant Pancreatic and Colorectal Cancer.
Cancer Res., 79, 2019
|
|
5BMW
 
 | | Crystal structure of T75V mutant of Triosephosphate isomerase from Plasmodium falciparum | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Bandyopadhyay, D, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2015-05-23 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Probing the role of highly conserved residues in triosephosphate isomerase - analysis of site specific mutants at positions 64 and 75 in the Plasmodial enzyme
Febs J., 282, 2015
|
|
5BN8
 
 | |
1KYT
 
 | | Crystal Structure of Thermoplasma acidophilum 0175 (APC014) | | Descriptor: | CALCIUM ION, hypothetical protein TA0175 | | Authors: | Kim, Y, Joachimiak, A, Edwards, A, Xu, X, Pennycooke, M, Gu, J, Cheung, F, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-02-05 | | Release date: | 2003-01-21 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Thermoplasma acidophilum 0175 (APC014)
To be published
|
|
6HYM
 
 | | Structure of PCM1 LIR motif bound to GABARAP | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Pericentriolar material 1 protein,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Mouilleron, S, Wirth, M, Zhang, W, O'Reilly, N, Tooze, S, Johansen, T, Razi, M, Nyoni, L, Joshi, D. | | Deposit date: | 2018-10-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Molecular determinants regulating selective binding of autophagy adapters and receptors to ATG8 proteins.
Nat Commun, 10, 2019
|
|
5BT3
 
 | | Crystal structure of EP300 bromodomain in complex with SGC-CBP30 chemical probe | | Descriptor: | 2-[2-(3-chloro-4-methoxyphenyl)ethyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(2S)-2-(morpholin-4-yl)propyl]-1H-benzimidazole, Histone acetyltransferase p300, ISOPROPYL ALCOHOL | | Authors: | Tallant, C, Hay, D, Krojer, T, Nunez-Alonso, G, Picaud, S, Newman, J.A, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-02 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Crystal structure of EP300 bromodomain in complex with a 3,5-dimethylisoxazol ligand
To Be Published
|
|
6HZO
 
 | | Apo structure of TP domain from Haemophilus influenzae Penicillin-Binding Protein 3 | | Descriptor: | FtsI | | Authors: | Bellini, D, Koekemoer, L, Newman, H, Dowson, C.G. | | Deposit date: | 2018-10-23 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Novel and Improved Crystal Structures of H. influenzae, E. coli and P. aeruginosa Penicillin-Binding Protein 3 (PBP3) and N. gonorrhoeae PBP2: Toward a Better Understanding of beta-Lactam Target-Mediated Resistance.
J.Mol.Biol., 431, 2019
|
|
6HZQ
 
 | | Apo structure of TP domain from Escherichia coli Penicillin-Binding Protein 3 | | Descriptor: | Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Bellini, D, Koekemoer, L, Newman, H, Dowson, C.G. | | Deposit date: | 2018-10-23 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel and Improved Crystal Structures of H. influenzae, E. coli and P. aeruginosa Penicillin-Binding Protein 3 (PBP3) and N. gonorrhoeae PBP2: Toward a Better Understanding of beta-Lactam Target-Mediated Resistance.
J.Mol.Biol., 431, 2019
|
|
5BU4
 
 | | RIBONUCLEASE T1 COMPLEX WITH 2'GMP | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, PROTEIN (RIBONUCLEASE T1) | | Authors: | Loris, R, Devos, S, Langhorst, U, Decanniere, K, Bouckaert, J, Maes, D, Transue, T.R, Steyaert, J. | | Deposit date: | 1998-09-15 | | Release date: | 1998-09-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Conserved water molecules in a large family of microbial ribonucleases.
Proteins, 36, 1999
|
|
1KZT
 
 | |
1KZZ
 
 | | DOWNSTREAM REGULATOR TANK BINDS TO THE CD40 RECOGNITION SITE ON TRAF3 | | Descriptor: | TNF receptor associated factor 3, TRAF family member-associated NF-kappa-b activator | | Authors: | Li, C, Ni, C.-Z, Havert, M.L, Cabezas, E, He, J, Kaiser, D, Reed, J.C, Satterthwait, A.C, Cheng, G, Ely, K.R. | | Deposit date: | 2002-02-08 | | Release date: | 2002-04-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Downstream regulator TANK binds to the CD40 recognition site on TRAF3.
Structure, 10, 2002
|
|
5BUU
 
 | | Crystal structure of the GluA2 ligand-binding domain (L483Y-N754S) in complex with glutamate and BPAM-321 at 2.07 A resolution | | Descriptor: | (3R)-7-chloro-2,3,4-trimethyl-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, 1,2-ETHANEDIOL, GLUTAMIC ACID, ... | | Authors: | Larsen, A.P, Tapken, D, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2015-06-04 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Synthesis and Pharmacology of Mono-, Di-, and Trialkyl-Substituted 7-Chloro-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-Dioxides Combined with X-ray Structure Analysis to Understand the Unexpected Structure-Activity Relationship at AMPA Receptors.
Acs Chem Neurosci, 7, 2016
|
|
5BUX
 
 | |
6HWR
 
 | | Red kidney bean purple acid phosphatase in complex with adenosine divanadate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feder, D, Gahan, L.R, McGeary, R.P, Guddat, L.W, Schenk, G. | | Deposit date: | 2018-10-13 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Binding Mode of an ADP Analogue to a Metallohydrolase Mimics the Likely Transition State.
Chembiochem, 20, 2019
|
|
5B5Z
 
 | | Crystal structure of PtLCIB4 H88A mutant, a homolog of the limiting CO2-inducible protein LCIB | | Descriptor: | PtLCIB4 H88A mutant, ZINC ION | | Authors: | Jin, S, Sun, J, Wunder, T, Tang, D, Mueller-Caja, O.M, Gao, Y. | | Deposit date: | 2016-05-24 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the LCIB protein family reveals a new group of beta-carbonic anhydrases
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
6HY3
 
 | | Three-dimensional structure of AgaC from Zobellia galactanivorans | | Descriptor: | 1,2-ETHANEDIOL, Beta-agarase C, GLYCEROL, ... | | Authors: | Naretto, A, Fanuel, M, Ropartz, D, Rogniaux, H, Larocque, R, Czjzek, M, Tellier, C, Michel, G. | | Deposit date: | 2018-10-19 | | Release date: | 2019-03-13 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The agar-specific hydrolaseZgAgaC from the marine bacteriumZobellia galactanivoransdefines a new GH16 protein subfamily.
J.Biol.Chem., 294, 2019
|
|
