1RZR
 
 | | crystal structure of transcriptional regulator-phosphoprotein-DNA complex | | Descriptor: | 5'-D(*CP*TP*GP*AP*AP*AP*GP*CP*GP*CP*TP*AP*AP*CP*AP*G)-3', 5'-D(*CP*TP*GP*TP*TP*AP*GP*CP*GP*CP*TP*TP*TP*CP*AP*G)-3', Glucose-resistance amylase regulator, ... | | Authors: | Schumacher, M.A, Allen, G.S, Brennan, R.G. | | Deposit date: | 2003-12-27 | | Release date: | 2004-10-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for allosteric control of the transcription regulator CcpA by the phosphoprotein HPr-Ser46-P.
Cell(Cambridge,Mass.), 118, 2004
|
|
1SXI
 
 | | Structure of apo transcription regulator B. megaterium | | Descriptor: | Glucose-resistance amylase regulator, MAGNESIUM ION | | Authors: | Schumacher, M.A, Allen, G.S, Diel, M, Seidel, G, Hillen, W, Brennan, R.G. | | Deposit date: | 2004-03-30 | | Release date: | 2004-10-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural studies on the apo transcription factor form B. megaterium
Cell(Cambridge,Mass.), 118, 2004
|
|
1SXG
 
 | | Structural studies on the apo transcription factor form B. megaterium | | Descriptor: | 2-PHENYLAMINO-ETHANESULFONIC ACID, Glucose-resistance amylase regulator | | Authors: | Schumacher, M.A, Allen, G.S, Diel, M, Seidel, G, Hillen, W, Brennan, R.G. | | Deposit date: | 2004-03-30 | | Release date: | 2004-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural studies on the apo transcription factor form B. megaterium
Cell(Cambridge,Mass.), 118, 2004
|
|
1SDK
 
 | | CROSS-LINKED, CARBONMONOXY HEMOGLOBIN A | | Descriptor: | 1,3,5-BENZENETRICARBOXYLIC ACID, CARBON MONOXIDE, HEMOGLOBIN A, ... | | Authors: | Schumacher, M.A, Dixon, M.M, Kluger, R, Jones, R.T, Brennan, R.G. | | Deposit date: | 1996-02-26 | | Release date: | 1996-08-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Allosteric transition intermediates modelled by crosslinked haemoglobins.
Nature, 375, 1995
|
|
1UPU
 
 | | STRUCTURE OF THE URACIL PHOSPHORIBOSYLTRANSFERASE, MUTANT C128V, BOUND TO PRODUCT URIDINE-1-MONOPHOSPHATE (UMP) | | Descriptor: | PHOSPHATE ION, URACIL PHOSPHORIBOSYLTRANSFERASE, URIDINE-5'-MONOPHOSPHATE | | Authors: | Schumacher, M.A, Carter, D, Scott, D, Roos, D, Ullman, B, Brennan, R.G. | | Deposit date: | 1998-04-16 | | Release date: | 1999-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Toxoplasma gondii uracil phosphoribosyltransferase reveal the atomic basis of pyrimidine discrimination and prodrug binding.
EMBO J., 17, 1998
|
|
1UPF
 
 | | STRUCTURE OF THE URACIL PHOSPHORIBOSYLTRANSFERASE, MUTANT C128V BOUND TO THE DRUG 5-FLUOROURACIL | | Descriptor: | 5-FLUOROURACIL, SULFATE ION, URACIL PHOSPHORIBOSYLTRANSFERASE | | Authors: | Schumacher, M.A, Carter, D, Scott, D, Roos, D, Ullman, B, Brennan, R.G. | | Deposit date: | 1998-06-17 | | Release date: | 1999-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of Toxoplasma gondii uracil phosphoribosyltransferase reveal the atomic basis of pyrimidine discrimination and prodrug binding.
EMBO J., 17, 1998
|
|
1SXH
 
 | | apo structure of B. megaterium transcription regulator | | Descriptor: | Glucose-resistance amylase regulator | | Authors: | Schumacher, M.A, Allen, G.S, Diel, M, Seidel, G, Hillen, W, Brennan, R.G. | | Deposit date: | 2004-03-30 | | Release date: | 2004-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural studies on the apo transcription factor form B. megaterium
Cell(Cambridge,Mass.), 118, 2004
|
|
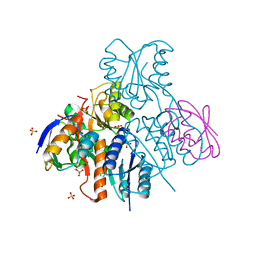
2NZU
 
 | | Structural mechanism for the fine-tuning of CcpA function by the small molecule effectors G6P and FBP | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, Catabolite control protein, Phosphocarrier protein HPr, ... | | Authors: | Schumacher, M.A, Hillen, W, Brennan, R.G. | | Deposit date: | 2006-11-25 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Mechanism for the Fine-tuning of CcpA Function by The Small Molecule Effectors Glucose 6-Phosphate and Fructose 1,6-Bisphosphate.
J.Mol.Biol., 368, 2007
|
|
2NTZ
 
 | |
2NZV
 
 | | Structural mechanism for the fine-tuning of CcpA function by the small molecule effectors G6P and FBP | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Catabolite control protein, Phosphocarrier protein HPr, ... | | Authors: | Schumacher, M.A, Hillen, W, Brennan, R.G. | | Deposit date: | 2006-11-25 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Mechanism for the Fine-tuning of CcpA Function by The Small Molecule Effectors Glucose 6-Phosphate and Fructose 1,6-Bisphosphate.
J.Mol.Biol., 368, 2007
|
|
2OEN
 
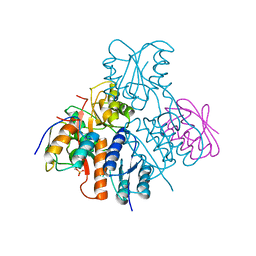 | | Structural mechanism for the fine-tuning of CcpA function by the small molecule effectors glucose-6-phosphate and fructose-1,6-bisphosphate | | Descriptor: | Catabolite control protein, Phosphocarrier protein HPr | | Authors: | Schumacher, M.A, Seidel, G, Hillen, W, Brennan, R.G. | | Deposit date: | 2006-12-30 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Structural Mechanism for the Fine-tuning of CcpA Function by The Small Molecule Effectors Glucose 6-Phosphate and Fructose 1,6-Bisphosphate.
J.Mol.Biol., 368, 2007
|
|
2Q2K
 
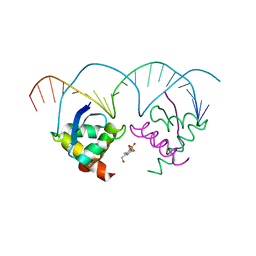 | | Structure of nucleic-acid binding protein | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DNA (5'-D(*AP*GP*TP*AP*TP*AP*(5IU)P*AP*CP*(5IU)P*AP*GP*TP*AP*TP*AP*TP*AP*CP*T)-3'), Hypothetical protein | | Authors: | Schumacher, M.A, Glover, T, Firth, N. | | Deposit date: | 2007-05-28 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Segrosome structure revealed by a complex of ParR with centromere DNA.
Nature, 450, 2007
|
|
3Q5W
 
 | |
3DNT
 
 | | structures of MDT proteins | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein hipA, ... | | Authors: | Schumacher, M.A. | | Deposit date: | 2008-07-02 | | Release date: | 2009-01-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB.
Science, 323, 2009
|
|
3DNV
 
 | | MDT Protein | | Descriptor: | DNA (5'-D(*DAP*DCP*DTP*DAP*DTP*DCP*DCP*DCP*DCP*DTP*DTP*DAP*DAP*DGP*DGP*DGP*DGP*DAP*DTP*DAP*DG)-3'), HTH-type transcriptional regulator hipB, Protein hipA, ... | | Authors: | schumacher, M.A. | | Deposit date: | 2008-07-02 | | Release date: | 2009-01-27 | | Last modified: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB.
Science, 323, 2009
|
|
3FBR
 
 | | structure of HipA-amppnp-peptide | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Serine/threonine-protein kinase toxin HipA, peptide of EF-Tu | | Authors: | Schumacher, M.A. | | Deposit date: | 2008-11-19 | | Release date: | 2009-02-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB.
Science, 323, 2009
|
|
3DNU
 
 | | structure of MDT protein | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Protein hipA | | Authors: | schumacher, M.A. | | Deposit date: | 2008-07-02 | | Release date: | 2009-01-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB.
Science, 323, 2009
|
|
5I41
 
 | | Structure of the apo RacA DNA binding domain | | Descriptor: | Chromosome-anchoring protein RacA | | Authors: | schumacher, M.A. | | Deposit date: | 2016-02-11 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular insights into DNA binding and anchoring by the Bacillus subtilis sporulation kinetochore-like RacA protein.
Nucleic Acids Res., 44, 2016
|
|
1PNR
 
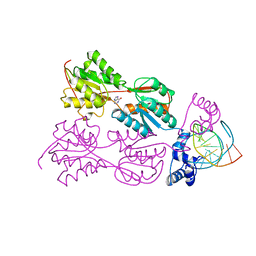 | | PURINE REPRESSOR-HYPOXANTHINE-PURF-OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*TP*TP*TP*TP*CP*GP*T )-3'), HYPOXANTHINE, PROTEIN (PURINE REPRESSOR) | | Authors: | Schumacher, M.A, Choi, K.Y, Zalkin, H, Brennan, R.G. | | Deposit date: | 1995-03-29 | | Release date: | 1995-11-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of LacI member, PurR, bound to DNA: minor groove binding by alpha helices.
Science, 266, 1994
|
|
4E07
 
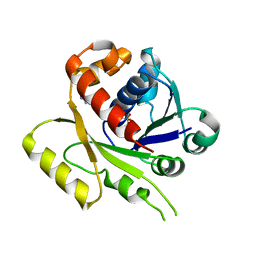 | | ParF-AMPPCP-C2221 form | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Plasmid partitioning protein ParF | | Authors: | Schumacher, M.A, Ye, Q, Barge, M.R, Barilla, D, Hayes, F. | | Deposit date: | 2012-03-02 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Mechanism of ATP-induced Polymerization of the Partition Factor ParF: IMPLICATIONS FOR DNA SEGREGATION.
J.Biol.Chem., 287, 2012
|
|
4E03
 
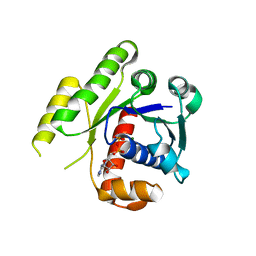 | | Structure of ParF-ADP form 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Plasmid partitioning protein ParF | | Authors: | Schumacher, M.A, Ye, Q, Barge, M.R, Barilla, D, Hayes, F. | | Deposit date: | 2012-03-02 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Mechanism of ATP-induced Polymerization of the Partition Factor ParF: IMPLICATIONS FOR DNA SEGREGATION.
J.Biol.Chem., 287, 2012
|
|
4DZZ
 
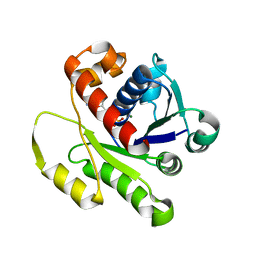 | | Structure of ParF-ADP, crystal form 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Plasmid partitioning protein ParF | | Authors: | Schumacher, M.A, Ye, Q, Barge, M.R, Barilla, D, Hayes, F. | | Deposit date: | 2012-03-01 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Mechanism of ATP-induced Polymerization of the Partition Factor ParF: IMPLICATIONS FOR DNA SEGREGATION.
J.Biol.Chem., 287, 2012
|
|
4E09
 
 | | Structure of ParF-AMPPCP, I422 form | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Plasmid partitioning protein ParF, SULFATE ION | | Authors: | Schumacher, M.A, Ye, Q, Barge, M.R, Barilla, D, Hayes, F. | | Deposit date: | 2012-03-02 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural Mechanism of ATP-induced Polymerization of the Partition Factor ParF: IMPLICATIONS FOR DNA SEGREGATION.
J.Biol.Chem., 287, 2012
|
|
1JUS
 
 | | Crystal structure of the multidrug binding transcriptional repressor QacR bound to rhodamine 6G | | Descriptor: | HYPOTHETICAL TRANSCRIPTIONAL REGULATOR IN QACA 5'REGION, RHODAMINE 6G, SULFATE ION | | Authors: | Schumacher, M.A, Miller, M.C, Grkovic, S, Brown, M.H, Skurray, R.A, Brennan, R.G. | | Deposit date: | 2001-08-27 | | Release date: | 2001-12-12 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structural mechanisms of QacR induction and multidrug recognition.
Science, 294, 2001
|
|
1JTY
 
 | | Crystal structure of the multidrug binding transcriptional regulator QacR bound to ethidium | | Descriptor: | ETHIDIUM, HYPOTHETICAL TRANSCRIPTIONAL REGULATOR IN QACA 5'REGION, SULFATE ION | | Authors: | Schumacher, M.A, Miller, M.C, Grkovic, S, Brown, M.H, Skurray, R.A, Brennan, R.G. | | Deposit date: | 2001-08-22 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structural mechanisms of QacR induction and multidrug recognition.
Science, 294, 2001
|
|
