7SGW
 
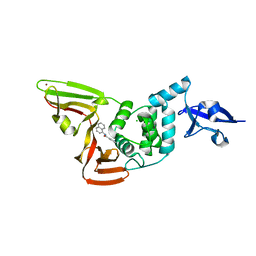 | | Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder630 inhibitor | | Descriptor: | CHLORIDE ION, N-(naphthalen-1-yl)pyridine-3-carboxamide, Non-structural protein 3, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-10-07 | | Release date: | 2021-10-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder630 inhibitor
To Be Published
|
|
2LY8
 
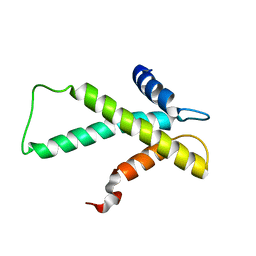 | | The budding yeast chaperone Scm3 recognizes the partially unfolded dimer of the centromere-specific Cse4/H4 histone variant | | Descriptor: | Budding yeast chaperone Scm3 | | Authors: | Hong, J, Feng, H, Zhou, Z, Ghirlando, R, Bai, Y. | | Deposit date: | 2012-09-13 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Identification of Functionally Conserved Regions in the Structure of the Chaperone/CenH3/H4 Complex.
J.Mol.Biol., 425, 2013
|
|
9JCV
 
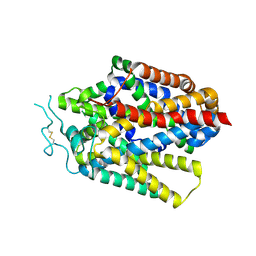 | | Cryo-EM structure of human TauT in the apo state, determined in an inward-facing open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Sodium- and chloride-dependent taurine transporter | | Authors: | Qu, Q, Chao, Y, Zhou, Z. | | Deposit date: | 2024-08-30 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Transport and inhibition mechanism for human TauT-mediated taurine uptake.
Cell Res., 35, 2025
|
|
9JD6
 
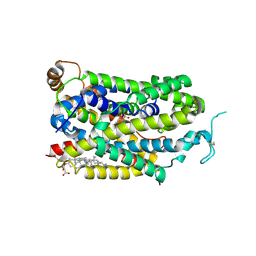 | | Cryo-EM structure of human TauT in presence of Piperidine-4-sulfonate, determined in an inward-facing occluded conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Qu, Q, Chao, Y, Zhou, Z. | | Deposit date: | 2024-08-30 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Transport and inhibition mechanism for human TauT-mediated taurine uptake.
Cell Res., 35, 2025
|
|
9JD5
 
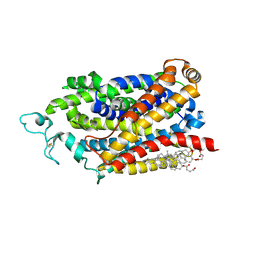 | | Cryo-EM structure of human TauT in presence of Taurocyamine, determined in an inward-facing occluded conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Qu, Q, Chao, Y, Zhou, Z. | | Deposit date: | 2024-08-30 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Transport and inhibition mechanism for human TauT-mediated taurine uptake.
Cell Res., 35, 2025
|
|
9JCZ
 
 | | Cryo-EM structure of human TauT in presence of taurine, determined in an inward-facing occluded conformation | | Descriptor: | 2-AMINOETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Qu, Q, Chao, Y, Zhou, Z. | | Deposit date: | 2024-08-30 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Transport and inhibition mechanism for human TauT-mediated taurine uptake.
Cell Res., 35, 2025
|
|
9JLN
 
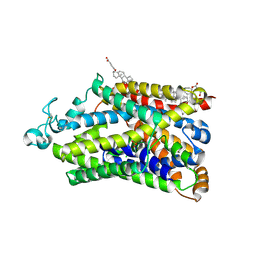 | | Cryo-EM structure of human TauT in presence of taurine, observed only one sodium ion, determined in an inward-occluded conformation | | Descriptor: | 2-AMINOETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Qu, Q, Chao, Y, Zhou, Z. | | Deposit date: | 2024-09-19 | | Release date: | 2025-03-19 | | Last modified: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Transport and inhibition mechanism for human TauT-mediated taurine uptake.
Cell Res., 35, 2025
|
|
8XN1
 
 | | Cryo-EM structure of human ZnT3, in the presence of zinc, determined in an inward-facing conformation | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, CHOLESTEROL HEMISUCCINATE, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Zhu, Z, Qu, Q, Long, Y, Zhou, Z. | | Deposit date: | 2023-12-28 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural insights into human zinc transporter ZnT1 mediated Zn 2+ efflux.
Embo Rep., 25, 2024
|
|
7KOK
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder496 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[(E)-(hydroxyimino)methyl]-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-09 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder496
to be published
|
|
8XMJ
 
 | | Cryo-EM structure of human ZnT1 WT, in the presence of zinc, determined in heterogeneous conformations- one subunit in an inward-facing and the other in an outward-facing conformation | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Lauryl Maltose Neopentyl Glycol, Proton-coupled zinc antiporter SLC30A1, ... | | Authors: | Qu, Q, Long, Y, Zhou, Z. | | Deposit date: | 2023-12-27 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Structural insights into human zinc transporter ZnT1 mediated Zn 2+ efflux.
Embo Rep., 25, 2024
|
|
8XM6
 
 | | Cryo-EM structure of human ZnT1 WT, in the absence of zinc, determined in an outward-facing conformation | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Lauryl Maltose Neopentyl Glycol, Proton-coupled zinc antiporter SLC30A1, ... | | Authors: | Qu, Q, Long, Y, Zhou, Z. | | Deposit date: | 2023-12-27 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural insights into human zinc transporter ZnT1 mediated Zn 2+ efflux.
Embo Rep., 25, 2024
|
|
8XMA
 
 | | Cryo-EM structure of human ZnT1 WT, in the presence of zinc, determined in an outward-facing conformation | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Lauryl Maltose Neopentyl Glycol, Proton-coupled zinc antiporter SLC30A1, ... | | Authors: | Qu, Q, Long, Y, Zhou, Z. | | Deposit date: | 2023-12-27 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structural insights into human zinc transporter ZnT1 mediated Zn 2+ efflux.
Embo Rep., 25, 2024
|
|
8XMF
 
 | | Cryo-EM structure of human ZnT1 WT at a low PH, in the presence of zinc, determined in an inward-facing conformation. | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Lauryl Maltose Neopentyl Glycol, Proton-coupled zinc antiporter SLC30A1, ... | | Authors: | Qu, Q, Long, Y, Zhou, Z. | | Deposit date: | 2023-12-27 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural insights into human zinc transporter ZnT1 mediated Zn 2+ efflux.
Embo Rep., 25, 2024
|
|
7KRX
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder441 inhibitor | | Descriptor: | 3-amino-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-20 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder441
to be published
|
|
7KOL
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder496 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[(E)-(hydroxyimino)methyl]-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-09 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder496
to be published
|
|
8WLJ
 
 | |
7DFG
 
 | | Structure of COVID-19 RNA-dependent RNA polymerase bound to favipiravir | | Descriptor: | 6-fluoro-3-oxo-4-(5-O-phosphono-beta-D-ribofuranosyl)-3,4-dihydropyrazine-2-carboxamide, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Li, Z, Zhou, Z, Yu, X. | | Deposit date: | 2020-11-08 | | Release date: | 2021-11-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for repurpose and design of nucleotide drugs for treating COVID-19
To Be Published
|
|
7DFH
 
 | |
7D4F
 
 | | Structure of COVID-19 RNA-dependent RNA polymerase bound to suramin | | Descriptor: | 8-(3-(3-aminobenzamido)-4-methylbenzamido)naphthalene-1,3,5-trisulfonic acid, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Li, Z, Yin, W, Zhou, Z, Yu, X, Xu, H. | | Deposit date: | 2020-09-23 | | Release date: | 2020-11-11 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 RNA polymerase by suramin.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6A7U
 
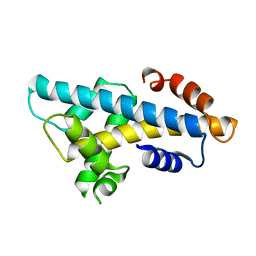 | | Crystal structure of histone H2A.Bbd-H2B dimer | | Descriptor: | Histone H2B type 2-E,Histone H2A-Bbd type 2/3 | | Authors: | Dai, L, Zhou, Z. | | Deposit date: | 2018-07-04 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the histone heterodimer containing histone variant H2A.Bbd.
Biochem. Biophys. Res. Commun., 503, 2018
|
|
7WLP
 
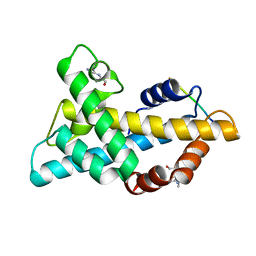 | |
6AE8
 
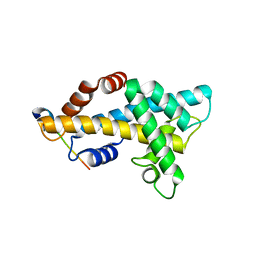 | | Structure insight into histone chaperone Chz1-mediated H2A.Z recognition and replacement | | Descriptor: | BICINE, Histone H2A.Z-specific chaperone CHZ1, Histone H2B.1,Histone H2A.Z | | Authors: | Wang, Y.Y, Shan, S, Zhou, Z. | | Deposit date: | 2018-08-03 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into histone chaperone Chz1-mediated H2A.Z recognition and histone replacement.
Plos Biol., 17, 2019
|
|
8Z8Z
 
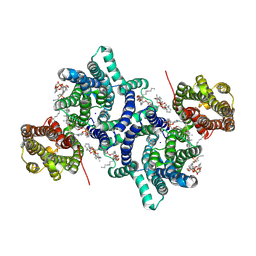 | | Cryo-EM structure of LYCHOS | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Xiong, Q, Zhu, Z, Li, T, Zhou, Z, Chao, Y, Qu, Q, Li, D. | | Deposit date: | 2024-04-22 | | Release date: | 2025-01-01 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.11 Å) | | Cite: | Molecular architecture of human LYCHOS involved in lysosomal cholesterol signaling.
Nat.Struct.Mol.Biol., 32, 2025
|
|
7X5B
 
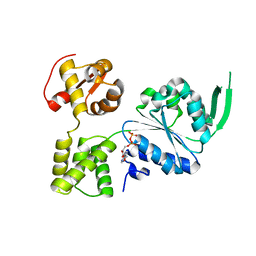 | | Crystal structure of RuvB | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvB | | Authors: | Lin, Z, Qu, Q, Zhang, X, Zhou, Z, Dai, L. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
7X5A
 
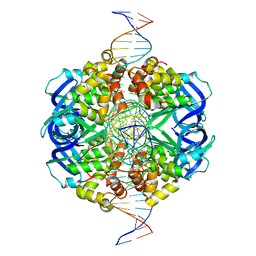 | | CryoEM structure of RuvA-Holliday junction complex | | Descriptor: | DNA (26-MER), Holliday junction ATP-dependent DNA helicase RuvA | | Authors: | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-08 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
