7XN9
 
 | | Crystal structure of SSTR2 and L-054,522 complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Somatostatin receptor type 2,Endo-1,4-beta-xylanase, tert-butyl (2S)-6-azanyl-2-[[(2R,3S)-3-(1H-indol-3-yl)-2-[[4-(2-oxidanylidene-3H-benzimidazol-1-yl)piperidin-1-yl]carbonylamino]butanoyl]amino]hexanoate | | Authors: | Zhao, W, Han, S, Qiu, N, Feng, W, Lu, M, Yang, D, Wang, M.-W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XMR
 
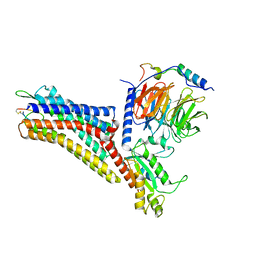 | | CryoEM structure of the somatostatin receptor 2 (SSTR2) in complex with Gi1 and its endogeneous peptide ligand SST-14 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wenli, Z, Shuo, H, Na, Q, Wenbo, Z, Mengjie, L, Dehua, Y, Ming-Wei, W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XMS
 
 | | CryoEM structure of somatostatin receptor 4 (SSTR4) in complex with Gi1 and its endogeneous ligand SST-14 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wenli, Z, Shuo, H, Na, Q, Wenbo, Z, Mengjie, L, Dehua, Y, Ming-Wei, W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XMT
 
 | | CryoEM structure of somatostatin receptor 4 (SSTR4) with Gi1 and J-2156 | | Descriptor: | (2~{S})-2-[[(2~{S})-4-azanyl-2-[(4-methylnaphthalen-1-yl)sulfonylamino]butanoyl]amino]-3-phenyl-propanimidic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wenli, Z, Shuo, H, Na, Q, Wenbo, Z, Mengjie, L, Dehua, Y, Ming-Wei, W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
3QV9
 
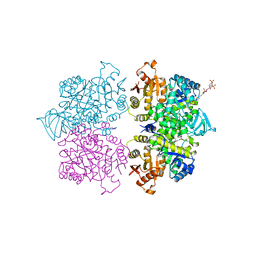 | | Crystal structure of Trypanosoma cruzi pyruvate kinase(TcPYK)in complex with ponceau S. | | Descriptor: | 3-hydroxy-4-[(E)-{2-sulfo-4-[(E)-(4-sulfophenyl)diazenyl]phenyl}diazenyl]naphthalene-2,7-disulfonic acid, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Morgan, H.P, Auld, D.S, McNae, I.W, Nowicki, M.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2011-02-25 | | Release date: | 2011-07-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The trypanocidal drug suramin and other trypan blue mimetics are inhibitors of pyruvate kinases and bind to the adenosine site.
J.Biol.Chem., 286, 2011
|
|
3QV7
 
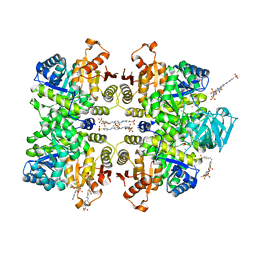 | | Crystal structure of Leishmania mexicana pyruvate kinase(LmPYK)in complex with ponceau S and acid blue 25. | | Descriptor: | 3-hydroxy-4-[(E)-{2-sulfo-4-[(E)-(4-sulfophenyl)diazenyl]phenyl}diazenyl]naphthalene-2,7-disulfonic acid, 9,10-dioxo-4-(phenylamino)-9,10-dihydroanthracene-2-sulfonic acid, POTASSIUM ION, ... | | Authors: | Morgan, H.P, Auld, D.S, McNae, I.W, Nowicki, M.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2011-02-25 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The trypanocidal drug suramin and other trypan blue mimetics are inhibitors of pyruvate kinases and bind to the adenosine site.
J.Biol.Chem., 286, 2011
|
|
3QV6
 
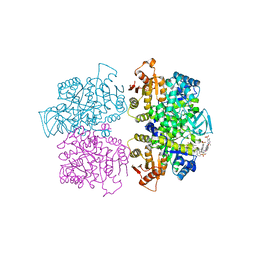 | | Crystal structure of Leishmania mexicana pyruvate kinase(LmPYK)in complex with acid blue 80. | | Descriptor: | 3-({4-[(2,4-dimethyl-5-sulfophenyl)amino]-9,10-dioxo-9,10-dihydroanthracen-1-yl}amino)-2,4,6-trimethylbenzenesulfonic acid, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Morgan, H.P, Auld, D.S, McNae, I.W, Nowicki, M.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2011-02-25 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The trypanocidal drug suramin and other trypan blue mimetics are inhibitors of pyruvate kinases and bind to the adenosine site.
J.Biol.Chem., 286, 2011
|
|
3QV8
 
 | | Crystal structure of Leishmania mexicana pyruvate kinase(LmPYK)in complex with benzothiazole-2,5-disulfonic acid. | | Descriptor: | 1,3-benzothiazole-2,5-disulfonic acid, PHOSPHATE ION, Pyruvate kinase | | Authors: | Morgan, H.P, Auld, D.S, McNae, I.W, Nowicki, M.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2011-02-25 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The trypanocidal drug suramin and other trypan blue mimetics are inhibitors of pyruvate kinases and bind to the adenosine site.
J.Biol.Chem., 286, 2011
|
|
3PP7
 
 | | Crystal structure of Leishmania mexicana pyruvate kinase in complex with the drug suramin, an inhibitor of glycolysis. | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, GLYCEROL, POTASSIUM ION, ... | | Authors: | Morgan, H.P, Auld, D.S, McNae, I.W, Nowicki, M.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2010-11-24 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The trypanocidal drug suramin and other trypan blue mimetics are inhibitors of pyruvate kinases and bind to the adenosine site.
J.Biol.Chem., 286, 2011
|
|
4DI2
 
 | | Crystal structure of BACE1 in complex with hydroxyethylamine inhibitor 37 | | Descriptor: | (2R)-N-{(2S,3R)-4-{[(4'S)-6'-(2,2-dimethylpropyl)-3',4'-dihydrospiro[cyclobutane-1,2'-pyrano[2,3-b]pyridin]-4'-yl]amino}-3-hydroxy-1-[3-(1,3-thiazol-2-yl)phenyl]butan-2-yl}-2-methoxypropanamide, Beta-secretase 1, GLYCEROL | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2012-01-30 | | Release date: | 2012-10-10 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and synthesis of potent, orally efficacious hydroxyethylamine derived beta-site amyloid precursor protein cleaving enzyme (BACE1) inhibitors.
J.Med.Chem., 55, 2012
|
|
4DUS
 
 | | Structure of Bace-1 (Beta-Secretase) in complex with N-((2S,3R)-1-(4-fluorophenyl)-3-hydroxy-4-((6'-neopentyl-3',4'-dihydrospiro[cyclobutane-1,2'-pyrano[2,3-b]pyridin]-4'-yl)amino)butan-2-yl)acetamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Sickmier, E.A. | | Deposit date: | 2012-02-22 | | Release date: | 2012-10-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Potent and Orally Efficacious, Hydroxyethylamine-Based Inhibitor of beta-Secretase.
ACS Med Chem Lett, 3, 2012
|
|
4DH6
 
 | | Structure of Bace-1 (Beta-Secretase) in Complex with (2R)-N-((2S,3R)-1-(benzo[d][1,3]dioxol-5-yl)-3-hydroxy-4-((S)-6'-neopentyl-3',4'-dihydrospiro[cyclobutane-1,2'-pyrano[2,3-b]pyridine]-4'-ylamino)butan-2-yl)-2-methoxypropanamide | | Descriptor: | (2R)-N-[(2S,3R)-1-(1,3-benzodioxol-5-yl)-4-{[(4'S)-6'-(2,2-dimethylpropyl)-3',4'-dihydrospiro[cyclobutane-1,2'-pyrano[2,3-b]pyridin]-4'-yl]amino}-3-hydroxybutan-2-yl]-2-methoxypropanamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Sickmier, E.A. | | Deposit date: | 2012-01-27 | | Release date: | 2012-04-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and preparation of a potent series of hydroxyethylamine containing beta-secretase inhibitors that demonstrate robust reduction of central beta-amyloid.
J.Med.Chem., 55, 2012
|
|
4FS4
 
 | | Structure of BACE Bound to (S)-4-(3'-methoxy-[1,1'-biphenyl]-3-yl)-1,4-dimethyl-6-oxotetrahydropyrimidin-2(1H)-iminium | | Descriptor: | (6S)-2-amino-6-(3'-methoxybiphenyl-3-yl)-3,6-dimethyl-5,6-dihydropyrimidin-4(3H)-one, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Stamford, A. | | Deposit date: | 2012-06-26 | | Release date: | 2012-10-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A Potent and Orally Efficacious, Hydroxyethylamine-Based Inhibitor of beta-Secretase.
ACS Med Chem Lett, 3, 2012
|
|
8XJJ
 
 | | Co-crystal structure of SOS-1 and a potent, selective and orally bioavailable SOS1 inhibitor RGT-018 | | Descriptor: | 1,2-ETHANEDIOL, 5-[4-[[(1~{R})-1-[3-[bis(fluoranyl)methyl]-2-fluoranyl-phenyl]ethyl]amino]-2-methyl-6-morpholin-4-yl-7-oxidanylidene-pyrido[4,3-d]pyrimidin-8-yl]pyridine-2-carbonitrile, Son of sevenless homolog 1 | | Authors: | Ren, X. | | Deposit date: | 2023-12-21 | | Release date: | 2024-08-14 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of RGT-018: A Potent, Selective, and Orally Bioavailable SOS1 Inhibitor for KRAS-Driven Cancers.
Mol.Cancer Ther., 23, 2024
|
|
5WL0
 
 | | Co-crystal structure of Influenza A H3N2 PB2 (241-741) bound to VX-787 | | Descriptor: | (2S,3S)-3-[[5-fluoranyl-2-(5-fluoranyl-1H-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-4-yl]amino]bicyclo[2.2.2]octane-2-carboxylic acid, DI(HYDROXYETHYL)ETHER, Polymerase basic protein 2 | | Authors: | Ma, X, Shia, S. | | Deposit date: | 2017-07-25 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for therapeutic inhibition of influenza A polymerase PB2 subunit.
Sci Rep, 7, 2017
|
|
4HF4
 
 | |
4HEU
 
 | |
4K9H
 
 | | Bace-1 inhibitor complex | | Descriptor: | 1-cyclopentyl-N-[(2S,3R)-3-hydroxy-1-phenyl-4-{[3-(trifluoromethyl)benzyl]amino}butan-2-yl]-6-oxo-5-(2-oxopyrrolidin-1-yl)-1,6-dihydropyridine-3-carboxamide, Beta-secretase 1 | | Authors: | Jordan, S.R. | | Deposit date: | 2013-04-19 | | Release date: | 2013-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Hydroxyethylamine-based inhibitors of BACE1: P1-P3 macrocyclization can improve potency, selectivity, and cell activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4K8S
 
 | | Hydroxyethylamine-based inhibitors of BACE1: P1-P3 macrocyclization can improve potency, selectivity, and cell activity | | Descriptor: | (3S)-3-[(1R)-2-{[(4S)-6-ethyl-3,4-dihydrospiro[chromene-2,1'-cyclobutan]-4-yl]amino}-1-hydroxyethyl]-4-azabicyclo[11.3.1]heptadeca-1(17),13,15-trien-5-one, Beta-secretase 1 | | Authors: | Jordan, S.R. | | Deposit date: | 2013-04-18 | | Release date: | 2013-07-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Hydroxyethylamine-based inhibitors of BACE1: P1-P3 macrocyclization can improve potency, selectivity, and cell activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4KE1
 
 | | Crystal structure of BACE1 in complex with hydroxyethylamine-macrocyclic inhibitor 19 | | Descriptor: | (12S)-12-[(1R)-2-{[(4S)-6-ethyl-3,4-dihydrospiro[chromene-2,1'-cyclobutan]-4-yl]amino}-1-hydroxyethyl]-1,13-diazatricyclo[13.3.1.1~6,10~]icosa-6(20),7,9,15(19),16-pentaene-14,18-dione, Beta-Secretase 1, GLYCEROL, ... | | Authors: | Whittington, D.A, Long, A.M, Li, V. | | Deposit date: | 2013-04-25 | | Release date: | 2013-07-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Hydroxyethylamine-based inhibitors of BACE1: P1-P3 macrocyclization can improve potency, selectivity, and cell activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4KE0
 
 | | Crystal structure of BACE1 in complex with hydroxyethylamine-macrocyclic inhibitor 13 | | Descriptor: | (3S)-3-[(1R)-2-{[(4S)-6-ethyl-3,4-dihydrospiro[chromene-2,1'-cyclobutan]-4-yl]amino}-1-hydroxyethyl]-4-azabicyclo[10.3.1]hexadeca-1(16),12,14-trien-5-one, Beta-secretase 1, GLYCEROL, ... | | Authors: | Whittington, D.A, Long, A.M, Li, V. | | Deposit date: | 2013-04-25 | | Release date: | 2013-07-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Hydroxyethylamine-based inhibitors of BACE1: P1-P3 macrocyclization can improve potency, selectivity, and cell activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5HZW
 
 | | Crystal structure of the orphan region of human endoglin/CD105 in complex with BMP9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Growth/differentiation factor 2, Maltose-binding periplasmic protein,Endoglin, ... | | Authors: | Bokhove, M, Saito, T, Jovine, L. | | Deposit date: | 2016-02-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.451 Å) | | Cite: | Structural Basis of the Human Endoglin-BMP9 Interaction: Insights into BMP Signaling and HHT1.
Cell Rep, 19, 2017
|
|
5I04
 
 | | Crystal structure of the orphan region of human endoglin/CD105 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Maltose-binding periplasmic protein,Endoglin, TRIETHYLENE GLYCOL, ... | | Authors: | Saito, T, Bokhove, M, de Sanctis, D, Jovine, L. | | Deposit date: | 2016-02-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Basis of the Human Endoglin-BMP9 Interaction: Insights into BMP Signaling and HHT1.
Cell Rep, 19, 2017
|
|
7XQL
 
 | | complex structure of LegA15 with GNP | | Descriptor: | Ankyrin repeat-containing protein, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Chen, T.T, Lin, Y.L. | | Deposit date: | 2022-05-07 | | Release date: | 2023-05-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.272 Å) | | Cite: | Atypical Legionella GTPase effector hijacks host vesicular transport factor p115 to regulate host lipid droplet.
Sci Adv, 8, 2022
|
|
7VQK
 
 | |
