6W0K
 
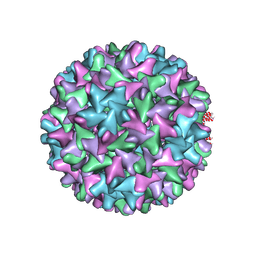 | | HBV D78S mutant capsid | | Descriptor: | Capsid protein | | Authors: | Zhao, Z, Wang, J, Zlotnick, A. | | Deposit date: | 2020-03-01 | | Release date: | 2020-09-30 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | The Integrity of the Intradimer Interface of the Hepatitis B Virus Capsid Protein Dimer Regulates Capsid Self-Assembly.
Acs Chem.Biol., 15, 2020
|
|
7XCP
 
 | | Cryo-EM structure of Omicron RBD complexed with ACE2 and 304 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 304 Fab, ... | | Authors: | Zhao, Z, Qi, J, Gao, F.G. | | Deposit date: | 2022-03-24 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
4N3X
 
 | |
4N3Z
 
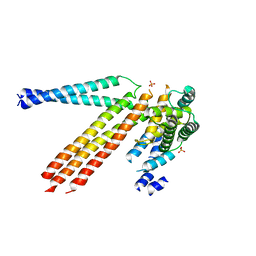 | |
4Q9U
 
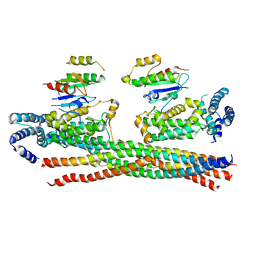 | | Crystal structure of the Rab5, Rabex-5delta and Rabaptin-5C21 complex | | Descriptor: | Rab GTPase-binding effector protein 1, Rab5 GDP/GTP exchange factor, Ras-related protein Rab-5A | | Authors: | Zhang, Z, Zhang, T, Ding, J. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.618 Å) | | Cite: | Molecular mechanism for Rabex-5 GEF activation by Rabaptin-5
Elife, 3, 2014
|
|
2BCC
 
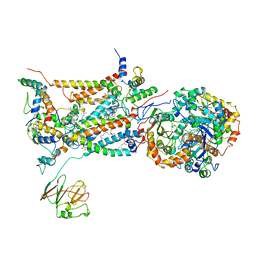 | | STIGMATELLIN-BOUND CYTOCHROME BC1 COMPLEX FROM CHICKEN | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, FE2/S2 (INORGANIC) CLUSTER, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.I, Kim, K.K, Hung, L.W, Crofts, A.R, Berry, E.A, Kim, S.H. | | Deposit date: | 1998-09-18 | | Release date: | 1999-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Electron Transfer by Domain Movement in Cytochrome Bc1
Nature, 392, 1998
|
|
4H20
 
 | | Crystal Structure and Computational Modeling of the Fab Fragment from the Protective anti-Ricin Monoclonal Antibody RAC18 | | Descriptor: | Heavy chain of Fab Fragment from the anti-Ricin Monoclonal Antibody RAC18, Light chain of Fab Fragment from the anti-Ricin Monoclonal Antibody RAC18 | | Authors: | Zhao, Z, Worthylake, D.K, LeCour Jr, L.F, Maresh, G, Pincus, S.H. | | Deposit date: | 2012-09-11 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and computational modeling of the fab fragment from a protective anti-ricin monoclonal antibody.
Plos One, 7, 2012
|
|
8UVK
 
 | | CosR DNA bound form II | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA (5'-D(*AP*AP*TP*TP*AP*AP*GP*AP*TP*AP*TP*TP*AP*TP*TP*AP*AP*CP*CP*AP*A)-3'), DNA (5'-D(*TP*TP*GP*GP*TP*TP*AP*AP*TP*AP*AP*TP*AP*TP*CP*TP*TP*AP*AP*TP*T)-3'), ... | | Authors: | Zhang, Z. | | Deposit date: | 2023-11-03 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural basis of DNA recognition of the Campylobacter jejuni CosR regulator.
Mbio, 15, 2024
|
|
8UUZ
 
 | | Campylobacter jejuni CosR apo form | | Descriptor: | DNA-binding response regulator | | Authors: | Zhang, Z. | | Deposit date: | 2023-11-02 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis of DNA recognition of the Campylobacter jejuni CosR regulator.
Mbio, 15, 2024
|
|
8UVX
 
 | | CosR DNA bound form I | | Descriptor: | DNA (5'-D(*AP*TP*AP*TP*CP*TP*TP*AP*AP*TP*TP*TP*TP*GP*GP*TP*TP*AP*AP*TP*A)-3'), DNA (5'-D(*TP*AP*TP*TP*AP*AP*CP*CP*AP*AP*AP*AP*TP*TP*AP*AP*GP*AP*TP*AP*T)-3'), DNA-binding response regulator | | Authors: | Zhang, Z. | | Deposit date: | 2023-11-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of DNA recognition of the Campylobacter jejuni CosR regulator.
Mbio, 15, 2024
|
|
3DA8
 
 | | Crystal structure of PurN from Mycobacterium tuberculosis | | Descriptor: | BETA-MERCAPTOETHANOL, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Zhang, Z, Squire, C.J, Baker, E.N. | | Deposit date: | 2008-05-28 | | Release date: | 2009-05-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of glycinamide ribonucleotide transformylase (PurN) from Mycobacterium tuberculosis reveal a novel dimer with relevance to drug discovery.
J.Mol.Biol., 389, 2009
|
|
3DCJ
 
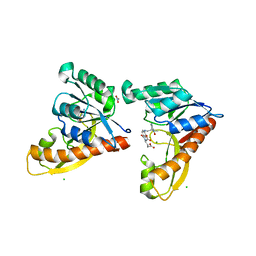 | | Crystal structure of glycinamide formyltransferase (PurN) from Mycobacterium tuberculosis in complex with 5-methyl-5,6,7,8-tetrahydrofolic acid derivative | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-[4-({[(6S)-2-AMINO-4-HYDROXY-5-METHYL-5,6,7,8-TETRAHYDROPTERIDIN-6-YL]METHYL}AMINO)BENZOYL]-L-GLUTAMIC ACID, ... | | Authors: | Zhang, Z, Squire, C.J, Baker, E.N. | | Deposit date: | 2008-06-03 | | Release date: | 2009-05-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of glycinamide ribonucleotide transformylase (PurN) from Mycobacterium tuberculosis reveal a novel dimer with relevance to drug discovery.
J.Mol.Biol., 389, 2009
|
|
3D98
 
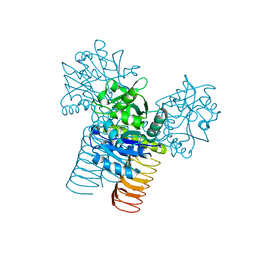 | |
3DZB
 
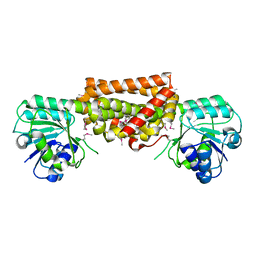 | |
1RW2
 
 | | Three-dimensional structure of Ku80 CTD | | Descriptor: | ATP-dependent DNA helicase II, 80 kDa subunit | | Authors: | Zhang, Z, Hu, W, Cano, L, Lee, T.D, Chen, D.J, Chen, Y. | | Deposit date: | 2003-12-15 | | Release date: | 2003-12-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal domain of Ku80 suggests important sites for protein-protein interactions.
STRUCTURE, 12, 2004
|
|
3BCC
 
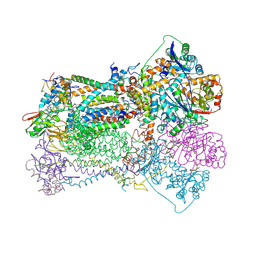 | | STIGMATELLIN AND ANTIMYCIN BOUND CYTOCHROME BC1 COMPLEX FROM CHICKEN | | Descriptor: | ANTIMYCIN, FE2/S2 (INORGANIC) CLUSTER, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, Z, Huang, L, Shulmeister, V.M, Chi, Y.-I, Kim, K.K, Hung, L.-W, Crofts, A.R, Berry, E.A, Kim, S.-H. | | Deposit date: | 1998-03-23 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Electron transfer by domain movement in cytochrome bc1.
Nature, 392, 1998
|
|
3EAF
 
 | | Crystal structure of ABC transporter, substrate binding protein Aeropyrum pernix | | Descriptor: | ABC transporter, substrate binding protein, GLYCEROL, ... | | Authors: | Zhang, Z, Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-25 | | Release date: | 2008-09-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of ABC transporter, substrate binding protein Aeropyrum pernix
To be Published
|
|
3FKD
 
 | |
3D8V
 
 | |
3EVZ
 
 | |
8DMP
 
 | |
8DMU
 
 | |
8DMS
 
 | |
8DMR
 
 | |
8DMQ
 
 | |
