2GC7
 
 | | Substrate reduced, copper free complex of methylamine dehydrogenase, amicyanin and cytochrome c551i from Paracoccus denitrificans. | | Descriptor: | Amicyanin, Cytochrome c-L, HEME C, ... | | Authors: | Chen, Z, Durley, R, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2006-03-13 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structral comparison of the oxidized ternary electron transfer complex of methylamine dehydrogenase, amicyanin and cytochrome c551i from Paracoccus denitrificans with the substrate-reduced, copper free complex at 1.9 A resolution.
To be Published
|
|
2WTM
 
 | | Est1E from Butyrivibrio proteoclasticus | | Descriptor: | EST1E, GLYCEROL, PHOSPHATE ION | | Authors: | Goldstone, D.C, Arcus, V.L. | | Deposit date: | 2009-09-17 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Characterization of a Promiscuous Feruloyl Esterase (Est1E) from the Rumen Bacterium Butyrivibrio Proteoclasticus.
Proteins, 78, 2010
|
|
2WTN
 
 | | Ferulic Acid bound to Est1E from Butyrivibrio proteoclasticus | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, EST1E, GLYCEROL, ... | | Authors: | Goldstone, D.C, Arcus, V.L. | | Deposit date: | 2009-09-17 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Functional Characterization of a Promiscuous Feruloyl Esterase (Est1E) from the Rumen Bacterium Butyrivibrio Proteoclasticus.
Proteins, 78, 2010
|
|
2XUT
 
 | | Crystal structure of a proton dependent oligopeptide (POT) family transporter. | | Descriptor: | PROTON/PEPTIDE SYMPORTER FAMILY PROTEIN | | Authors: | Newstead, S, Drew, D, Cameron, A.D, Postis, V.L, Xia, X, Fowler, P.W, Ingram, J.C, Carpenter, E.P, Sansom, M.S.P, McPherson, M.J, Baldwin, S.A, Iwata, S. | | Deposit date: | 2010-10-21 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Crystal Structure of a Prokaryotic Homologue of the Mammalian Oligopeptide-Proton Symporters, Pept1 and Pept2.
Embo J., 30, 2011
|
|
2XB4
 
 | | Crystal structures of zinc containing Adenylate kinase from Desulfovibrio gigas | | Descriptor: | ADENYLATE KINASE, S,R MESO-TARTARIC ACID, ZINC ION | | Authors: | Mukhopadhyay, A, Kladova, A.V, Gavel, O.Y, Calvete, J.J, Shnyrov, V.L, Moura, I, Moura, J.J.G, Bursakov, S.A, Romao, M.J, Trincao, J. | | Deposit date: | 2010-04-05 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Zinc-, Cobalt-, and Iron-Containing Adenylate Kinase from Desulfovibrio Gigas: A Novel Metal-Containing Adenylate Kinase from Gram-Negative Bacteria.
J.Biol.Inorg.Chem., 16, 2011
|
|
8BLU
 
 | | The PDZ domains of human SDCBP | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ALANINE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Daniel-Mozo, M, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2022-11-10 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The PDZ domains of human SDCBP
To Be Published
|
|
8BNT
 
 | | The DH domain of ARHGEF2 bound to RhoA | | Descriptor: | DIMETHYL SULFOXIDE, FORMIC ACID, Rho guanine nucleotide exchange factor 2, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Grosjean, H, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2022-11-25 | | Release date: | 2022-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The DH domain of ARHGEF2 bound to RhoA
To Be Published
|
|
8BLV
 
 | | The PDZ domains of human SDCBP with a bound SDC4 C-terminal peptide | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Syndecan-4, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Daniel-Mozo, M, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2022-11-10 | | Release date: | 2022-12-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The PDZ domains of human SDCBP with a bound SDC4 C-terminal peptide
To Be Published
|
|
8CIS
 
 | | The FERM domain of human moesin with two bound peptides identified by phage display | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, C3P, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Leisner, T.M, Fairhead, M, Bountra, C, von Delft, F, Pearce, K.H, Brennan, P.E. | | Deposit date: | 2023-02-10 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of FERM domain protein-protein interaction inhibitors for MSN and CD44 as a potential therapeutic approach for Alzheimer's disease.
J.Biol.Chem., 299, 2023
|
|
8CIU
 
 | | The FERM domain of human moesin mutant H288A | | Descriptor: | Moesin | | Authors: | Bradshaw, W.J, Katis, V.L, Koekemoer, L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-02-10 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Discovery of FERM domain protein-protein interaction inhibitors for MSN and CD44 as a potential therapeutic approach for Alzheimer's disease.
J.Biol.Chem., 299, 2023
|
|
8CIT
 
 | | The FERM domain of human moesin mutant L281R | | Descriptor: | Moesin | | Authors: | Bradshaw, W.J, Katis, V.L, Koekemoer, L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-02-10 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.536 Å) | | Cite: | Discovery of FERM domain protein-protein interaction inhibitors for MSN and CD44 as a potential therapeutic approach for Alzheimer's disease.
J.Biol.Chem., 299, 2023
|
|
8CIR
 
 | | The FERM domain of human moesin with a bound peptide identified by phage display | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BROMIDE ION, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Leisner, T.M, Fairhead, M, Bountra, C, von Delft, F, Pearce, K.H, Brennan, P.E. | | Deposit date: | 2023-02-10 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of FERM domain protein-protein interaction inhibitors for MSN and CD44 as a potential therapeutic approach for Alzheimer's disease.
J.Biol.Chem., 299, 2023
|
|
3BGS
 
 | | Structure of human purine nucleoside phosphorylase with L-DADMe-ImmH and phosphate | | Descriptor: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, purine nucleoside phosphorylase | | Authors: | Murkin, A.S, Ramagopal, U.A, Almo, S.C, Schramm, V.L. | | Deposit date: | 2007-11-27 | | Release date: | 2008-01-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | L-Enantiomers of transition state analogue inhibitors bound to human purine nucleoside phosphorylase
J.Am.Chem.Soc., 130, 2008
|
|
2YAL
 
 | | SinR, Master Regulator of biofilm formation in Bacillus subtilis | | Descriptor: | HTH-TYPE TRANSCRIPTIONAL REGULATOR SINR, NICKEL (II) ION | | Authors: | Colledge, V.L, Fogg, M.J, Levdikov, V.M, Leech, A, Dodson, E.J, Wilkinson, A.J. | | Deposit date: | 2011-02-23 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure and Organisation of Sinr, the Master Regulator of Biofilm Formation in Bacillus Subtilis.
J.Mol.Biol., 411, 2011
|
|
2X6P
 
 | | Crystal Structure of Coil Ser L19C | | Descriptor: | COIL SER L19C, ZINC ION | | Authors: | Chakraborty, S, Touw, D.S, Peacock, A.F.A, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2010-02-18 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Comparisons of Apo- and Metalated Three-Stranded Coiled Coils Clarify Metal Binding Determinants in Thiolate Containing Designed Peptides.
J.Am.Chem.Soc., 132, 2010
|
|
3D4B
 
 | | Crystal structure of Sir2Tm in complex with Acetyl p53 peptide and DADMe-NAD+ | | Descriptor: | 5'-O-[(R)-{[(R)-{[(3R,4R)-1-(3-carbamoylbenzyl)-4-hydroxypyrrolidin-3-yl]methoxy}(hydroxy)phosphoryl]methyl}(hydroxy)phosphoryl]adenosine, Acetyl P53 peptide, NAD-dependent deacetylase, ... | | Authors: | Hawse, W.F, Hoff, K.G, Fatkins, D, Daines, A, Zubkova, O.V, Schramm, V.L, Zheng, W, Wolberger, C. | | Deposit date: | 2008-05-14 | | Release date: | 2008-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into intermediate steps in the Sir2 deacetylation reaction.
Structure, 16, 2008
|
|
3CMZ
 
 | | TEM-1 Class-A beta-lactamase L201P mutant apo structure | | Descriptor: | Beta-lactamase TEM, PHOSPHATE ION | | Authors: | Marciano, D.C, Wang, X, Wang, J, Chen, Y, Thomas, V.L, Shoichet, B.K, Palzkill, T. | | Deposit date: | 2008-03-24 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Genetic and structural characterization of an L201P global suppressor substitution in TEM-1 beta-lactamase
J.Mol.Biol., 384, 2008
|
|
3D81
 
 | | Sir2-S-alkylamidate complex crystal structure | | Descriptor: | NAD-dependent deacetylase, S-alkylamidate intermediate, ZINC ION | | Authors: | Hawse, W.F, Hoff, K.G, Fatkins, D, Daines, A, Zubkova, O.V, Schramm, V.L, Zheng, W, Wolberger, C. | | Deposit date: | 2008-05-22 | | Release date: | 2008-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into intermediate steps in the Sir2 deacetylation reaction.
Structure, 16, 2008
|
|
4W9B
 
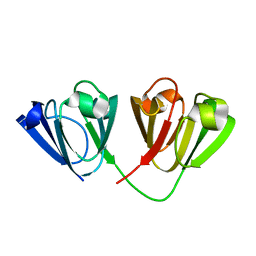 | | Crystal structure of Gamma-B Crystallin expressed in E. coli based on mRNA variant 1 | | Descriptor: | Gamma-crystallin B | | Authors: | Kudlinzki, D, Buhr, F, Linhard, V.L, Jha, S, Komar, A.A, Schwalbe, H. | | Deposit date: | 2014-08-27 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.279 Å) | | Cite: | Two synonymous gene variants encode proteins with identical sequence, but different folding conformations.
To Be Published
|
|
4WIH
 
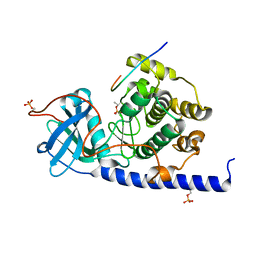 | | Crystal structure of cAMP-dependent Protein Kinase A from Cricetulus griseus | | Descriptor: | cAMP Dependent Protein Kinase Inhibitor PKI-tide, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Kudlinzki, D, Linhard, V.L, Saxena, K, Dreyer, M, Schwalbe, H. | | Deposit date: | 2014-09-25 | | Release date: | 2014-10-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.139 Å) | | Cite: | High-resolution crystal structure of cAMP-dependent protein kinase from Cricetulus griseus.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
1WTU
 
 | | TRANSCRIPTION FACTOR 1, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | TRANSCRIPTION FACTOR 1 | | Authors: | Jia, X, Grove, A, Ivancic, M, Hsu, V.L, Geiduschek, E.P, Kearns, D.R. | | Deposit date: | 1996-07-29 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Bacillus subtilis phage SPO1-encoded type II DNA-binding protein TF1 in solution.
J.Mol.Biol., 263, 1996
|
|
1Y6R
 
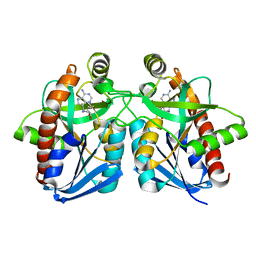 | | Crystal structure of MTA/AdoHcy nucleosidase complexed with MT-ImmA. | | Descriptor: | (3S,4R)-2-(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-5-[(METHYLSULFANYL)METHYL]PYRROLIDINE-3,4-DIOL, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Singh, V, Evans, G.B, Tyler, P.C, Furneaux, R.H, Cornell, K.A, Riscoe, M.K, Schramm, V.L, Howell, P.L. | | Deposit date: | 2004-12-06 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural rationale for the affinity of pico- and femtomolar transition state analogues of Escherichia coli 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase.
J.Biol.Chem., 280, 2005
|
|
2MAS
 
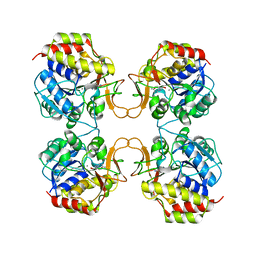 | | PURINE NUCLEOSIDE HYDROLASE WITH A TRANSITION STATE INHIBITOR | | Descriptor: | 2-(4-AMINO-PHENYL)-5-HYDROXYMETHYL-PYRROLIDINE-3,4-DIOL, CALCIUM ION, INOSINE-URIDINE NUCLEOSIDE N-RIBOHYDROLASE | | Authors: | Degano, M, Schramm, V.L, Sacchettini, J.C. | | Deposit date: | 1996-10-17 | | Release date: | 1997-08-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Trypanosomal nucleoside hydrolase. A novel mechanism from the structure with a transition-state inhibitor.
Biochemistry, 37, 1998
|
|
2PAC
 
 | | SOLUTION STRUCTURE OF FE(II) CYTOCHROME C551 FROM PSEUDOMONAS AERUGINOSA AS DETERMINED BY TWO-DIMENSIONAL 1H NMR | | Descriptor: | CYTOCHROME C551, HEME C | | Authors: | Detlefsen, D.J, Thanabal, V, Pecoraro, V.L, Wagner, G. | | Deposit date: | 1993-05-05 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Fe(II) cytochrome c551 from Pseudomonas aeruginosa as determined by two-dimensional 1H NMR.
Biochemistry, 30, 1991
|
|
5EUJ
 
 | | PYRUVATE DECARBOXYLASE FROM ZYMOBACTER PALMAE | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Pyruvate decarboxylase, ... | | Authors: | Buddrus, L, Crennell, S.J, Leak, D.J, Danson, M.J, Andrews, E.S.V, Arcus, V.L. | | Deposit date: | 2015-11-18 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of pyruvate decarboxylase from Zymobacter palmae.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
