8TM2
 
 | |
8TLZ
 
 | |
8VM1
 
 | |
3MDS
 
 | | MAGANESE SUPEROXIDE DISMUTASE FROM THERMUS THERMOPHILUS | | Descriptor: | MANGANESE (III) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Ludwig, M.L, Metzger, A.L, Pattridge, K.A, Stallings, W.C. | | Deposit date: | 1993-10-20 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Manganese superoxide dismutase from Thermus thermophilus. A structural model refined at 1.8 A resolution.
J.Mol.Biol., 219, 1991
|
|
4LRN
 
 | |
7SJQ
 
 | |
1B3J
 
 | | STRUCTURE OF THE MHC CLASS I HOMOLOG MIC-A, A GAMMADELTA T CELL LIGAND | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MHC CLASS I HOMOLOG MIC-A | | Authors: | Li, P, Willie, S, Bauer, S, Morris, D, Spies, T, Strong, R. | | Deposit date: | 1998-12-11 | | Release date: | 1999-07-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the MHC class I homolog MIC-A, a gammadelta T cell ligand.
Immunity, 10, 1999
|
|
1MNG
 
 | | STRUCTURE-FUNCTION IN E. COLI IRON SUPEROXIDE DISMUTASE: COMPARISONS WITH THE MANGANESE ENZYME FROM T. THERMOPHILUS | | Descriptor: | AZIDE ION, MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Lah, M.S, Dixon, M, Pattridge, K.A, Stallings, W.C, Fee, J.A, Ludwig, M.L. | | Deposit date: | 1994-07-13 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function in Escherichia coli iron superoxide dismutase: comparisons with the manganese enzyme from Thermus thermophilus.
Biochemistry, 34, 1995
|
|
4ODX
 
 | |
4OB5
 
 | |
6OL5
 
 | |
6OL6
 
 | | Structure of iglb12 scFv in complex with anti-idiotype ib2 Fab | | Descriptor: | CHLORIDE ION, ib2 Heavy Chain, ib2 Light Chain, ... | | Authors: | Weidle, C, Pancera, M, Gewe, M. | | Deposit date: | 2019-04-15 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Detection and activation of HIV broadly neutralizing antibody precursor B cells using anti-idiotypes.
J.Exp.Med., 216, 2019
|
|
7U61
 
 | | Crystal Structure of Anti-Nicotine Antibody NIC311 Fab Complexed with Nicotine | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, NIC311 Fab Heavy Chain, NIC311 Fab Light Chain, ... | | Authors: | Rodarte, J.V, Pancera, M.P, Liban, T.L. | | Deposit date: | 2022-03-03 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of drug-specific monoclonal antibodies bound to opioids and nicotine reveal a common mode of binding.
Structure, 31, 2023
|
|
7U62
 
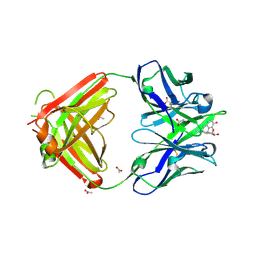 | | Crystal structure of Anti-Heroin Antibody HY4-1F9 Fab Complexed with Morphine | | Descriptor: | (7R,7AS,12BS)-3-METHYL-2,3,4,4A,7,7A-HEXAHYDRO-1H-4,12-METHANO[1]BENZOFURO[3,2-E]ISOQUINOLINE-7,9-DIOL, ACETATE ION, HY4-1F9 Fab Heavy Chain, ... | | Authors: | Rodarte, J.V, Pancera, M.P. | | Deposit date: | 2022-03-03 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structures of drug-specific monoclonal antibodies bound to opioids and nicotine reveal a common mode of binding.
Structure, 31, 2023
|
|
7U64
 
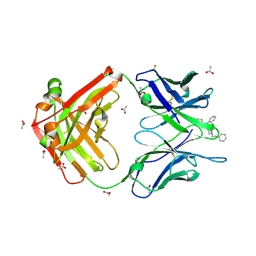 | |
3LRR
 
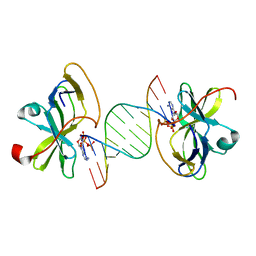 | | Crystal structure of human RIG-I CTD bound to a 12 bp AU rich 5' ppp dsRNA | | Descriptor: | Probable ATP-dependent RNA helicase DDX58, RNA (5'-R(*(ATP)P*UP*AP*UP*AP*UP*AP*UP*AP*UP*AP*U)-3'), ZINC ION | | Authors: | Li, P. | | Deposit date: | 2010-02-11 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Structural Basis of 5' Triphosphate Double-Stranded RNA Recognition by RIG-I C-Terminal Domain.
Structure, 18, 2010
|
|
3LEF
 
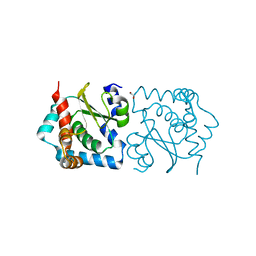 | | Crystal structure of HIV epitope-scaffold 4E10_S0_1Z6NA_001 | | Descriptor: | 1,2-ETHANEDIOL, Uncharacterized protein 4E10_S0_1Z6NA_001 (T18) | | Authors: | Holmes, M.A. | | Deposit date: | 2010-01-14 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Computational Design of Epitope-Scaffolds Allows Induction of Antibodies Specific for a Poorly Immunogenic HIV Vaccine Epitope.
Structure, 18, 2010
|
|
3LF6
 
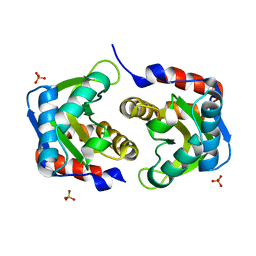 | | Crystal structure of HIV epitope-scaffold 4E10_1XIZA_S0_001_N | | Descriptor: | PHOSPHATE ION, Putative phosphotransferase system | | Authors: | Holmes, M.A. | | Deposit date: | 2010-01-15 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computational Design of Epitope-Scaffolds Allows Induction of Antibodies Specific for a Poorly Immunogenic HIV Vaccine Epitope.
Structure, 18, 2010
|
|
3LG7
 
 | | Crystal structure of HIV epitope-scaffold 4E10_S0_1EZ3A_002_C | | Descriptor: | 4E10_S0_1EZ3A_002_C (T246), SULFATE ION | | Authors: | Holmes, M.A. | | Deposit date: | 2010-01-19 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Computational Design of Epitope-Scaffolds Allows Induction of Antibodies Specific for a Poorly Immunogenic HIV Vaccine Epitope.
Structure, 18, 2010
|
|
3LHP
 
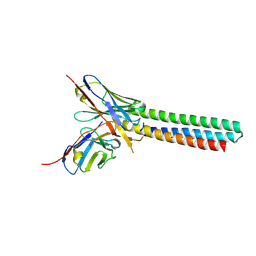 | |
3LH2
 
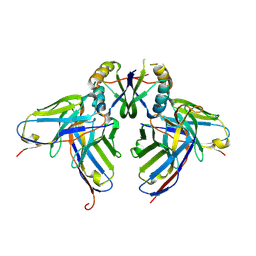 | |
3LF9
 
 | | Crystal structure of HIV epitope-scaffold 4E10_D0_1IS1A_001_C | | Descriptor: | 4E10_D0_1IS1A_001_C (T161) | | Authors: | Holmes, M.A. | | Deposit date: | 2010-01-16 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational Design of Epitope-Scaffolds Allows Induction of Antibodies Specific for a Poorly Immunogenic HIV Vaccine Epitope.
Structure, 18, 2010
|
|
3LRN
 
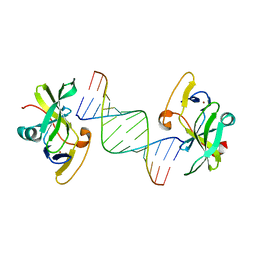 | | Crystal structure of human RIG-I CTD bound to a 14 bp GC 5' ppp dsRNA | | Descriptor: | Probable ATP-dependent RNA helicase DDX58, RNA (5'-R(*(GTP)P*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*C)-3'), ZINC ION | | Authors: | Li, P. | | Deposit date: | 2010-02-11 | | Release date: | 2010-06-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structural Basis of 5' Triphosphate Double-Stranded RNA Recognition by RIG-I C-Terminal Domain.
Structure, 18, 2010
|
|
7SQP
 
 | |
7SRK
 
 | |
