1A32
 
 | |
5MRC
 
 | | Structure of the yeast mitochondrial ribosome - Class A | | Descriptor: | 15S ribosomal RNA, 21S ribosomal RNA, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Desai, N, Brown, A, Amunts, A, Ramakrishnan, V. | | Deposit date: | 2016-12-22 | | Release date: | 2017-02-15 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | The structure of the yeast mitochondrial ribosome.
Science, 355, 2017
|
|
5MDZ
 
 | | Structure of the 70S ribosome (empty A site) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | James, N.R, Brown, A, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2016-11-13 | | Release date: | 2016-12-14 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Translational termination without a stop codon.
Science, 354, 2016
|
|
5OOL
 
 | | Structure of a native assembly intermediate of the human mitochondrial ribosome with unfolded interfacial rRNA | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Brown, A, Rathore, S, Kimanius, D, Aibara, S, Bai, X.C, Rorbach, J, Amunts, A, Ramakrishnan, V. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-13 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structures of the human mitochondrial ribosome in native states of assembly.
Nat. Struct. Mol. Biol., 24, 2017
|
|
4AQY
 
 | | Structure of ribosome-apramycin complexes | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Matt, T, Ng, C.L, Lang, K, Sha, S.H, Akbergenov, R, Shcherbakov, D, Meyer, M, Duscha, S, Xie, J, Dubbaka, S.R, Perez-Fernandez, D, Vasella, A, Ramakrishnan, V, Schacht, J, Bottger, E.C. | | Deposit date: | 2012-04-20 | | Release date: | 2012-07-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Dissociation of Antibacterial Activity and Aminoglycoside Ototoxicity in the 4-Monosubstituted 2-Deoxystreptamine Apramycin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5OOM
 
 | | Structure of a native assembly intermediate of the human mitochondrial ribosome with unfolded interfacial rRNA | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Brown, A, Rathore, S, Kimanius, D, Aibara, S, Bai, X.C, Rorbach, J, Amunts, A, Ramakrishnan, V. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-13 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structures of the human mitochondrial ribosome in native states of assembly.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5MRE
 
 | | Structure of the yeast mitochondrial ribosome - Class B | | Descriptor: | 15S ribosomal RNA, 21S ribosomal RNA, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Desai, N, Brown, A, Amunts, A, Ramakrishnan, V. | | Deposit date: | 2016-12-22 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | The structure of the yeast mitochondrial ribosome.
Science, 355, 2017
|
|
4B3T
 
 | | Crystal structure of the 30S ribosome in complex with compound 39 | | Descriptor: | (2S,3S,4R,5R,6R)-2-(aminomethyl)-5-azanyl-6-[(2R,3S,4R,5S)-5-[(1R,2R,3S,5R,6S)-3,5-bis(azanyl)-2-[(2S,3R,4R,5S,6R)-3-azanyl-5-[(4-chlorophenyl)methoxy]-6-(hydroxymethyl)-4-oxidanyl-oxan-2-yl]oxy-6-oxidanyl-cyclohexyl]oxy-2-(hydroxymethyl)-4-oxidanyl-oxolan-3-yl]oxy-oxane-3,4-diol, 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Shcherbakov, D, Matt, T, Perez-Fernandez, D, Patak, R, Meyer, M, Duscha, S, Akbergenov, R, Boukari, H, Freihofer, P, Kudyba, I, Reddy, M.S.K, Nandurikar, R.S, Ramakrishnan, V, Vasella, A, Bottger, E.C. | | Deposit date: | 2012-07-26 | | Release date: | 2013-08-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 4'-O-Substitutions Determine Selectivity of Aminoglycoside Antibiotics
Nat.Commun., 5, 2014
|
|
1BOB
 
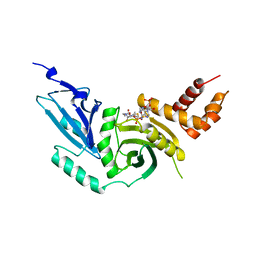 | | HISTONE ACETYLTRANSFERASE HAT1 FROM SACCHAROMYCES CEREVISIAE IN COMPLEX WITH ACETYL COENZYME A | | Descriptor: | ACETYL COENZYME *A, CALCIUM ION, HISTONE ACETYLTRANSFERASE | | Authors: | Dutnall, R.N, Tafrov, S.T, Sternglanz, R, Ramakrishnan, V. | | Deposit date: | 1998-07-02 | | Release date: | 1999-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the histone acetyltransferase Hat1: a paradigm for the GCN5-related N-acetyltransferase superfamily.
Cell(Cambridge,Mass.), 94, 1998
|
|
6QG1
 
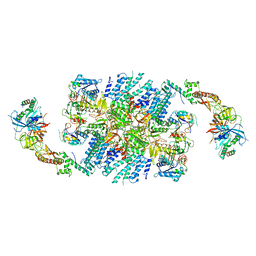 | | Structure of eIF2B-eIF2 (phosphorylated at Ser51) complex (model 2) | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, Eukaryotic translation initiation factor 2 subunit beta, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Llacer, J.L, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Structural basis for the inhibition of translation through eIF2 alpha phosphorylation.
Nat Commun, 10, 2019
|
|
6ZMW
 
 | | Structure of a human 48S translational initiation complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | Deposit date: | 2020-07-04 | | Release date: | 2020-09-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
6GSM
 
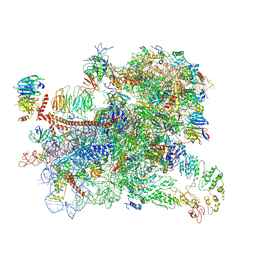 | | Structure of a partial yeast 48S preinitiation complex in open conformation. | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0, 40S ribosomal protein S1, ... | | Authors: | Llacer, J.L, Hussain, T, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2018-06-14 | | Release date: | 2019-07-31 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (5.15 Å) | | Cite: | Large-scale movement of eIF3 domains during translation initiation modulate start codon selection.
Nucleic Acids Res., 2021
|
|
6QG6
 
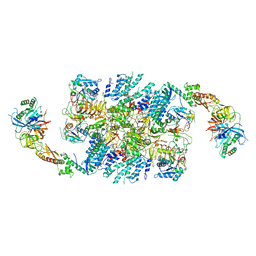 | | Structure of eIF2B-eIF2 (phosphorylated at Ser51) complex (model D) | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, Eukaryotic translation initiation factor 2 subunit beta, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Llacer, J.L, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (10.4 Å) | | Cite: | Structural basis for the inhibition of translation through eIF2 alpha phosphorylation.
Nat Commun, 10, 2019
|
|
6QG2
 
 | | Structure of eIF2B-eIF2 (phosphorylated at Ser51) complex (model A) | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, Eukaryotic translation initiation factor 2 subunit beta, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Llacer, J.L, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | Structural basis for the inhibition of translation through eIF2 alpha phosphorylation.
Nat Commun, 10, 2019
|
|
6QG0
 
 | | Structure of eIF2B-eIF2 (phosphorylated at Ser51) complex (model 1) | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, Eukaryotic translation initiation factor 2 subunit beta, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Llacer, J.L, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structural basis for the inhibition of translation through eIF2 alpha phosphorylation.
Nat Commun, 10, 2019
|
|
6QG3
 
 | | Structure of eIF2B-eIF2 (phosphorylated at Ser51) complex (model B) | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, Eukaryotic translation initiation factor 2 subunit beta, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Llacer, J.L, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Structural basis for the inhibition of translation through eIF2 alpha phosphorylation.
Nat Commun, 10, 2019
|
|
6QG5
 
 | | Structure of eIF2B-eIF2 (phosphorylated at Ser51) complex (model C) | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, Eukaryotic translation initiation factor 2 subunit beta, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Llacer, J.L, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (10.1 Å) | | Cite: | Structural basis for the inhibition of translation through eIF2 alpha phosphorylation.
Nat Commun, 10, 2019
|
|
1XNQ
 
 | |
1XMQ
 
 | | Crystal Structure of t6A37-ASLLysUUU AAA-mRNA Bound to the Decoding Center | | Descriptor: | 16s ribosomal RNA, 30S Ribosomal Protein S10, 30S Ribosomal Protein S11, ... | | Authors: | Murphy, F.V, Ramakrishnan, V, Malkiewicz, A, Agris, P.F. | | Deposit date: | 2004-10-04 | | Release date: | 2004-12-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The role of modifications in codon discrimination by tRNA(Lys)(UUU).
Nat.Struct.Mol.Biol., 11, 2004
|
|
1XNR
 
 | |
1XMO
 
 | | Crystal Structure of mnm5U34t6A37-tRNALysUUU Complexed with AAG-mRNA in the Decoding Center | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Murphy, F.V, Ramakrishnan, V, Malkiewicz, A, Agris, P.F. | | Deposit date: | 2004-10-04 | | Release date: | 2004-12-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The role of modifications in codon discrimination by tRNA(Lys)(UUU).
Nat.Struct.Mol.Biol., 11, 2004
|
|
4JYA
 
 | | Crystal structures of pseudouridinilated stop codons with ASLs | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Fernandez, I.S, Ng, C.L, Kelley, A.C, Guowei, W, Yu, Y.T, Ramakrishnan, V. | | Deposit date: | 2013-03-29 | | Release date: | 2013-06-26 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.098 Å) | | Cite: | Unusual base pairing during the decoding of a stop codon by the ribosome.
Nature, 500, 2013
|
|
4JV5
 
 | | Crystal structures of pseudouridinilated stop codons with ASLs | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein 20, 30S ribosomal protein S10, ... | | Authors: | Fernandez, I.S, Ng, C.L, Kelley, A.C, Guowei, W, Yu, Y.T, Ramakrishnan, V. | | Deposit date: | 2013-03-25 | | Release date: | 2013-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.162 Å) | | Cite: | Unusual base pairing during the decoding of a stop codon by the ribosome.
Nature, 500, 2013
|
|
4K0K
 
 | | Crystal structure of the Thermus thermophilus 30S ribosomal subunit complexed with a serine-ASL and mRNA containing a stop codon | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Fernandez, I.S, Ng, C.L, Kelley, A.C, Guowei, W, Yu, Y.T, Ramakrishnan, V. | | Deposit date: | 2013-04-04 | | Release date: | 2013-06-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Unusual base pairing during the decoding of a stop codon by the ribosome.
Nature, 500, 2013
|
|
4KHP
 
 | | Structure of the Thermus thermophilus 30S ribosomal subunit in complex with de-6-MSA-pactamycin | | Descriptor: | 16S Ribosomal RNA, 30S Ribosomal protein S10, 30S Ribosomal protein S11, ... | | Authors: | Tourigny, D.S, Fernandez, I.S, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2013-05-01 | | Release date: | 2013-06-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of a Bioactive Pactamycin Analog Bound to the 30S Ribosomal Subunit.
J.Mol.Biol., 425, 2013
|
|
