6IVZ
 
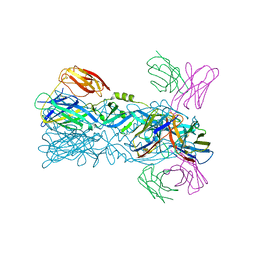 | | Crystal structure of 5A ScFv complexed with YFV-China sE in postfusion state | | Descriptor: | Envelope protein, Heavy chain of monoclonal antibody 5A, Light chain of monoclonal antibody 5A | | Authors: | Lu, X.S, Xiao, H.X, Li, S.H, Pang, X.F. | | Deposit date: | 2018-12-04 | | Release date: | 2019-02-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Double Lock of a Human Neutralizing and Protective Monoclonal Antibody Targeting the Yellow Fever Virus Envelope.
Cell Rep, 26, 2019
|
|
6IW0
 
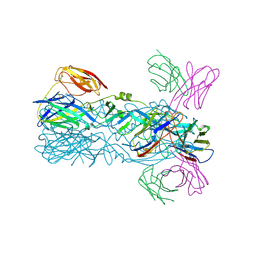 | | Crystal structure of 5A ScFv in complex with YFV-17D sE in postfusion state | | Descriptor: | Envelope protein E, Heavy chain of monoclonal antibody 5A, Light chain of monoclonal antibody 5A | | Authors: | Lu, X.S, Xiao, H.X, Li, S.H, Pang, X.F. | | Deposit date: | 2018-12-04 | | Release date: | 2019-02-13 | | Last modified: | 2019-02-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Double Lock of a Human Neutralizing and Protective Monoclonal Antibody Targeting the Yellow Fever Virus Envelope.
Cell Rep, 26, 2019
|
|
6IW2
 
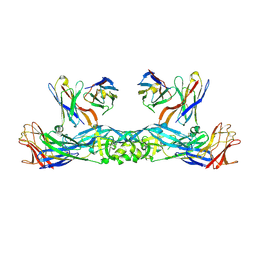 | | Crystal structure of 5A ScFv in complex with YFV-17D sE in prefusion state | | Descriptor: | Envelope protein E, Heavy chain of monoclonal antibody 5A, Light chain of monoclonal antibody 5A | | Authors: | Lu, X.S, Xiao, H.X, Li, S.H, Pang, X.F. | | Deposit date: | 2018-12-04 | | Release date: | 2019-02-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Double Lock of a Human Neutralizing and Protective Monoclonal Antibody Targeting the Yellow Fever Virus Envelope.
Cell Rep, 26, 2019
|
|
7EA6
 
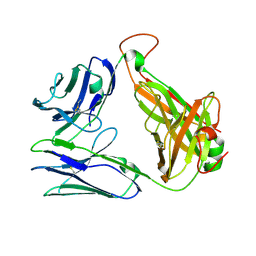 | | Crystal structure of TCR-017 ectodomain | | Descriptor: | T cell receptor 017 alpha chain, T cell receptor 017 beta chain | | Authors: | Nagae, M, Yamasaki, S. | | Deposit date: | 2021-03-06 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.18000245 Å) | | Cite: | Identification of conserved SARS-CoV-2 spike epitopes that expand public cTfh clonotypes in mild COVID-19 patients.
J.Exp.Med., 218, 2021
|
|
7DMX
 
 | |
7DNB
 
 | | Crystal structure of PhoCl barrel | | Descriptor: | PhoCl Barrel, SODIUM ION | | Authors: | Wen, Y, Lemieux, J.M. | | Deposit date: | 2020-12-09 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Photocleavable proteins that undergo fast and efficient dissociation.
Chem Sci, 12, 2021
|
|
7DNA
 
 | |
5HM4
 
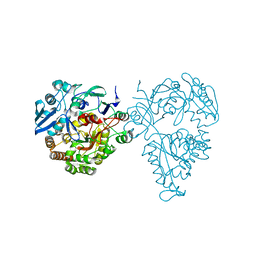 | | Crystal structure of oligopeptide ABC transporter, periplasmic oligopeptide-binding protein (TM1226) from THERMOTOGA MARITIMA at 2.0 A resolution | | Descriptor: | CALCIUM ION, Mannoside ABC transport system, sugar-binding protein | | Authors: | Lu, X, Ghimire-Rijal, S, Myles, D.A.A, Cuneo, M.J. | | Deposit date: | 2016-01-15 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Periplasmic Binding Protein Dimer Has a Second Allosteric Event Tied to Ligand Binding.
Biochemistry, 56, 2017
|
|
6E6J
 
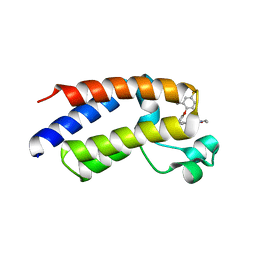 | | BRD2_Bromodomain2 complex with inhibitor 744 | | Descriptor: | Bromodomain-containing protein 2, N-ethyl-4-[2-(4-fluoro-2,6-dimethylphenoxy)-5-(2-hydroxypropan-2-yl)phenyl]-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide | | Authors: | Longenecker, K.L, Park, C.H, Bigelow, L. | | Deposit date: | 2018-07-25 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Selective inhibition of the BD2 bromodomain of BET proteins in prostate cancer.
Nature, 578, 2020
|
|
5X0S
 
 | | Solution NMR structure of peptide toxin SsTx from Scolopendra subspinipes mutilans | | Descriptor: | SsTx | | Authors: | Wu, F, Luo, L, Qu, D, Zhang, L, Tian, C, Lai, R. | | Deposit date: | 2017-01-23 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Centipedes subdue giant prey by blocking KCNQ channels
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1HNI
 
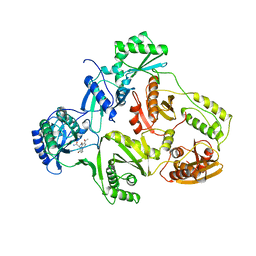 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN A COMPLEX WITH THE NONNUCLEOSIDE INHIBITOR ALPHA-APA R 95845 AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | (2-ACETYL-5-METHYLANILINO)(2,6-DIBROMOPHENYL)ACETAMIDE, HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66) | | Authors: | Ding, J, Das, K, Arnold, E. | | Deposit date: | 1995-02-28 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of HIV-1 reverse transcriptase in a complex with the non-nucleoside inhibitor alpha-APA R 95845 at 2.8 A resolution.
Structure, 3, 1995
|
|
1HNV
 
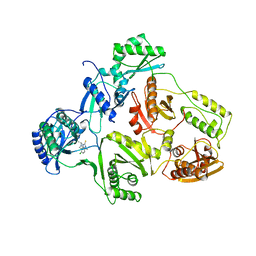 | | STRUCTURE OF HIV-1 RT(SLASH)TIBO R 86183 COMPLEX REVEALS SIMILARITY IN THE BINDING OF DIVERSE NONNUCLEOSIDE INHIBITORS | | Descriptor: | 5-CHLORO-8-METHYL-7-(3-METHYL-BUT-2-ENYL)-6,7,8,9-TETRAHYDRO-2H-2,7,9A-TRIAZA-BENZO[CD]AZULENE-1-THIONE, HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66) | | Authors: | Das, K, Ding, J, Arnold, E. | | Deposit date: | 1995-03-30 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of HIV-1 RT/TIBO R 86183 complex reveals similarity in the binding of diverse nonnucleoside inhibitors.
Nat.Struct.Biol., 2, 1995
|
|
3AL4
 
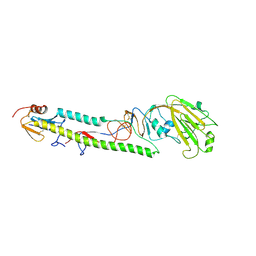 | | Crystal structure of the swine-origin A (H1N1)-2009 influenza A virus hemagglutinin (HA) reveals similar antigenicity to that of the 1918 pandemic virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Zhang, W, Qi, J.X, Shi, Y, Li, Q, Yan, J.H, Gao, G.F. | | Deposit date: | 2010-07-22 | | Release date: | 2010-08-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.872 Å) | | Cite: | Crystal structure of the swine-origin A (H1N1)-2009 influenza A virus hemagglutinin (HA) reveals similar antigenicity to that of the 1918 pandemic virus
Protein Cell, 1, 2010
|
|
6KZD
 
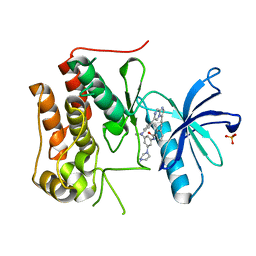 | | Crystal structure of TRKc in complex with 3-((6-(4-aminophenyl)imidazo[1,2-a]pyrazin-3-yl)ethynyl)- N-(3-isopropyl-5-((4-methylpiperazin-1-yl)methyl)phenyl)-2- methylbenzamide | | Descriptor: | 3-[2-[6-(4-aminophenyl)imidazo[1,2-a]pyrazin-3-yl]ethynyl]-2-methyl-~{N}-[3-(4-methylpiperazin-1-yl)-5-propan-2-yl-phenyl]benzamide, NT-3 growth factor receptor, PHOSPHATE ION | | Authors: | Wang, Y, Zhang, Z.M. | | Deposit date: | 2019-09-23 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.708 Å) | | Cite: | Design, synthesis and biological evaluation of 3-(imidazo[1,2-a]pyrazin-3-ylethynyl)-2-methylbenzamides as potent and selective pan-tropomyosin receptor kinase (TRK) inhibitors.
Eur.J.Med.Chem., 179, 2019
|
|
4RC6
 
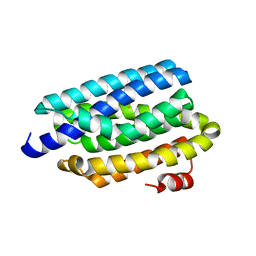 | | Crystal structure of cyanobacterial aldehyde-deformylating oxygenase 122F mutant | | Descriptor: | Aldehyde decarbonylase, FE (II) ION | | Authors: | Jia, C.J, Li, M, Li, J.J, Zhang, J.J, Zhang, H.M, Cao, P, Pan, X.W, Lu, X.F, Chang, W.R. | | Deposit date: | 2014-09-14 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the catalytic mechanism of aldehyde-deformylating oxygenases.
Protein Cell, 6, 2015
|
|
6GWR
 
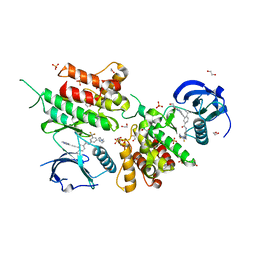 | | Structure of the kinase domain of human DDR1 in complex with a potent and selective inhibitor of DDR1 and DDR2 | | Descriptor: | 1,2-ETHANEDIOL, 3-(2-imidazo[1,2-a]pyrazin-3-ylethynyl)-~{N}-[3-[(4-methylpiperazin-1-yl)methyl]-5-(trifluoromethyl)phenyl]-4-propan-2-yl-benzamide, Epithelial discoidin domain-containing receptor 1, ... | | Authors: | Pinkas, D.M, Fox, A.E, Kupinska, K, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-06-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 3-(Imidazo[1,2- a]pyrazin-3-ylethynyl)-4-isopropyl- N-(3-((4-methylpiperazin-1-yl)methyl)-5-(trifluoromethyl)phenyl)benzamide as a Dual Inhibitor of Discoidin Domain Receptors 1 and 2.
J. Med. Chem., 61, 2018
|
|
6ONY
 
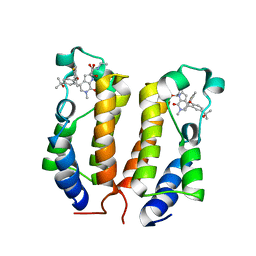 | | BRD2_Bromodomain1 complex with inhibitor 744 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 2, N-ethyl-4-[2-(4-fluoro-2,6-dimethylphenoxy)-5-(2-hydroxypropan-2-yl)phenyl]-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide | | Authors: | Longenecker, K.L, Bigelow, L. | | Deposit date: | 2019-04-22 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Selective inhibition of the BD2 bromodomain of BET proteins in prostate cancer.
Nature, 578, 2020
|
|
8YJY
 
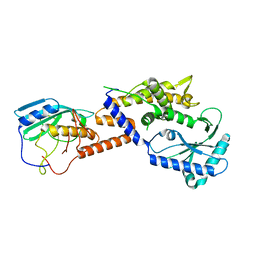 | |
8XZ7
 
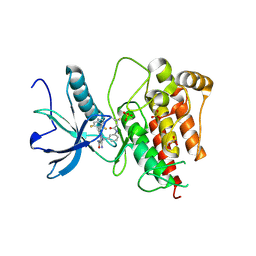 | | FGFR1 kinase domain with a covalent inhibitor 10h | | Descriptor: | 5-azanyl-3-[2-[4,6-bis(fluoranyl)-2-methyl-3~{H}-benzimidazol-5-yl]ethynyl]-1-[[3-(prop-2-enoylamino)phenyl]methyl]pyrazole-4-carboxamide, Fibroblast growth factor receptor 1, SULFATE ION | | Authors: | Chen, X.J, Chen, Y.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design, synthesis and biological evaluation of 5-amino-1H-pyrazole-4-carboxamide derivatives as pan-FGFR covalent inhibitors.
Eur.J.Med.Chem., 275, 2024
|
|
8Y22
 
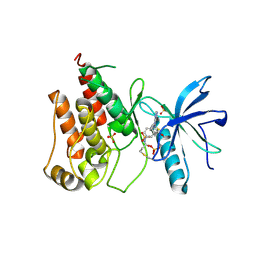 | | FGFR1 kinase domain with a covalent inhibitor 9g | | Descriptor: | Fibroblast growth factor receptor 1, SULFATE ION, ~{N}-[4-[[4-azanyl-3-(7-methoxy-5-methyl-1-benzothiophen-2-yl)pyrazolo[3,4-d]pyrimidin-1-yl]methyl]phenyl]propanamide | | Authors: | Chen, X.J, Chen, Y.H. | | Deposit date: | 2024-01-25 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.792 Å) | | Cite: | Design, synthesis and biological evaluation of 5-amino-1H-pyrazole-4-carboxamide derivatives as pan-FGFR covalent inhibitors.
Eur.J.Med.Chem., 275, 2024
|
|
7RR4
 
 | |
7RR3
 
 | | Structure of Deep-Sea Phage NrS-1 Primase-Polymerase N300 in complex with calcium and ddCTP | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, CALCIUM ION, Primase | | Authors: | Wang, L, Yu, C, Sliz, P. | | Deposit date: | 2021-08-09 | | Release date: | 2022-01-12 | | Last modified: | 2022-01-19 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Molecular Dissection of the Primase and Polymerase Activities of Deep-Sea Phage NrS-1 Primase-Polymerase.
Front Microbiol, 12, 2021
|
|
6OVG
 
 | |
8ZUD
 
 | | Crystal Structure of Human Myt1 Kinase domain Bounded with compound 8f | | Descriptor: | 2-azanyl-3-(2,6-dimethyl-3-oxidanyl-phenyl)-5-(2-morpholin-4-ylpyridin-4-yl)benzamide, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase | | Authors: | Zhang, Z.M, Zhou, Z.Q. | | Deposit date: | 2024-06-08 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.50510085 Å) | | Cite: | Structure-Based Drug Design of 2-Amino-[1,1'-biphenyl]-3-carboxamide Derivatives as Selective PKMYT1 Inhibitors for the Treatment of CCNE1 -Amplified Breast Cancer.
J.Med.Chem., 67, 2024
|
|
8ZTX
 
 | | Crystal Structure of Human Myt1 Kinase domain Bounded with compound 6b | | Descriptor: | 2-azanyl-3-(2,6-dimethyl-3-oxidanyl-phenyl)-5-pyridin-4-yl-benzamide, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase | | Authors: | Zhang, Z.M, Zhou, Z.Q. | | Deposit date: | 2024-06-07 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.70033228 Å) | | Cite: | Structure-Based Drug Design of 2-Amino-[1,1'-biphenyl]-3-carboxamide Derivatives as Selective PKMYT1 Inhibitors for the Treatment of CCNE1 -Amplified Breast Cancer.
J.Med.Chem., 67, 2024
|
|
