3TG4
 
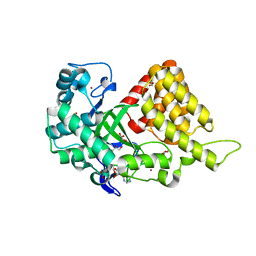 | | Structure of SMYD2 in complex with SAM | | Descriptor: | GLYCEROL, N-lysine methyltransferase SMYD2, S-ADENOSYLMETHIONINE, ... | | Authors: | Zhao, K, Wang, L. | | Deposit date: | 2011-08-17 | | Release date: | 2011-08-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human SMYD2 reveals the basis of p53 tumor suppressor methylation
J.Biol.Chem., 2011
|
|
5YAN
 
 | |
8SVF
 
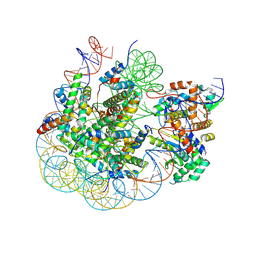 | | BAP1/ASXL1 bound to the H2AK119Ub Nucleosome | | Descriptor: | DNA/RNA (187-MER), DNA/RNA (327-MER), Histone H2A type 1, ... | | Authors: | Thomas, J.F, Valencia-Sanchez, M.I, Armache, K.-J. | | Deposit date: | 2023-05-16 | | Release date: | 2023-08-30 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of histone H2A lysine 119 deubiquitination by Polycomb repressive deubiquitinase BAP1/ASXL1.
Sci Adv, 9, 2023
|
|
8T9F
 
 | |
8T9H
 
 | |
5ZM6
 
 | | Crystal structure of ORP1-ORD in complex with PI(4,5)P2 | | Descriptor: | ACETATE ION, Oxysterol-binding protein-related protein 1, [(2~{S})-1-octadecanoyloxy-3-[oxidanyl-[(1~{R},2~{R},3~{S},4~{S},5~{S},6~{S})-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propan-2-yl] icosa-5,8,11,14-tetraenoate | | Authors: | Dong, J, Wang, J, Luo, Z, Wu, J.W. | | Deposit date: | 2018-04-01 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Allosteric enhancement of ORP1-mediated cholesterol transport by PI(4,5)P2/PI(3,4)P2.
Nat Commun, 10, 2019
|
|
5ZM7
 
 | | Crystal structure of ORP1-ORD in complex with cholesterol at 3.4 A resolution | | Descriptor: | CHOLESTEROL, Oxysterol-binding protein-related protein 1 | | Authors: | Dong, J, Wang, J, Wu, J.W. | | Deposit date: | 2018-04-01 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | Allosteric enhancement of ORP1-mediated cholesterol transport by PI(4,5)P2/PI(3,4)P2.
Nat Commun, 10, 2019
|
|
8THU
 
 | |
5ZM5
 
 | | Crystal structure of human ORP1-ORD in complex with cholesterol at 2.6 A resolution | | Descriptor: | CHOLESTEROL, Oxysterol-binding protein-related protein 1 | | Authors: | Dong, J, Wang, J, Wu, J.W. | | Deposit date: | 2018-04-01 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Allosteric enhancement of ORP1-mediated cholesterol transport by PI(4,5)P2/PI(3,4)P2.
Nat Commun, 10, 2019
|
|
6DBS
 
 | | Fusion surface structure, function, and dynamics of gamete fusogen HAP2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hapless 2, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feng, J, Dong, X, Springer, T.A. | | Deposit date: | 2018-05-03 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Fusion surface structure, function, and dynamics of gamete fusogen HAP2.
Elife, 7, 2018
|
|
7VHR
 
 | | Apostichopus japonicus ferritin | | Descriptor: | Ferritin, MAGNESIUM ION | | Authors: | Wu, Y, Su, X.R, Ming, T.H. | | Deposit date: | 2021-09-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.756 Å) | | Cite: | Crystallographic characterization of a marine invertebrate ferritin from the sea cucumber Apostichopus japonicus.
Febs Open Bio, 12, 2022
|
|
4OR9
 
 | | Crystal structure of human calcineurin | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, FE (III) ION, ... | | Authors: | Li, S.J, Wang, J, Wu, J.W, Wang, Z.X. | | Deposit date: | 2014-02-11 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Cooperative autoinhibition and multi-level activation mechanisms of calcineurin
To be Published
|
|
4ORA
 
 | | Crystal structure of a human calcineurin mutant | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, FE (III) ION, ... | | Authors: | Li, S.J, Wang, J, Wu, J.W, Wang, Z.X. | | Deposit date: | 2014-02-11 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.747 Å) | | Cite: | Cooperative autoinhibition and multi-level activation mechanisms of calcineurin
To be Published
|
|
4ORB
 
 | | Crystal structure of mouse calcineurin | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, FE (III) ION, ... | | Authors: | Ma, L, Li, S.J, Wang, J, Wu, J.W, Wang, Z.X. | | Deposit date: | 2014-02-11 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.108 Å) | | Cite: | Cooperative autoinhibition and multi-level activation mechanisms of calcineurin
To be Published
|
|
4ORC
 
 | | Crystal structure of mammalian calcineurin | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, FE (III) ION, ... | | Authors: | Ma, L, Li, S.J, Wang, J, Wu, J.W, Wang, Z.X. | | Deposit date: | 2014-02-11 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cooperative autoinhibition and multi-level activation mechanisms of calcineurin
To be Published
|
|
5H14
 
 | | EED in complex with an allosteric PRC2 inhibitor EED666 | | Descriptor: | 2-[3-(3,5-dimethylpyrazol-1-yl)-4-nitro-phenyl]-3,4-dihydro-1H-isoquinoline, GLYCEROL, Histone-lysine N-methyltransferase EZH2, ... | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Molecular Basis of a Diverse Set of Polycomb Repressive Complex 2 Inhibitors Recognition by EED
PLoS ONE, 12, 2017
|
|
5H15
 
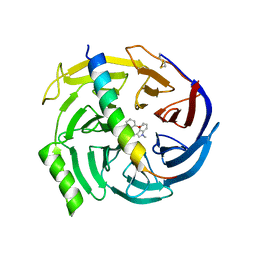 | | EED in complex with PRC2 allosteric inhibitor EED709 | | Descriptor: | (3R,4S)-1-[(2-methoxyphenyl)methyl]-N,N-dimethyl-4-(1-methylindol-3-yl)pyrrolidin-3-amine, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery and Molecular Basis of a Diverse Set of Polycomb Repressive Complex 2 Inhibitors Recognition by EED
PLoS ONE, 12, 2017
|
|
5FFG
 
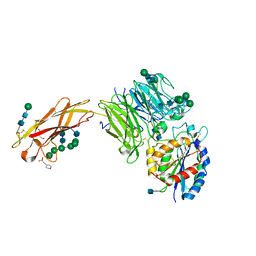 | | Crystal structure of integrin alpha V beta 6 head | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dong, X, Springer, T.A. | | Deposit date: | 2015-12-18 | | Release date: | 2017-01-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Force interacts with macromolecular structure in activation of TGF-beta.
Nature, 542, 2017
|
|
5FFO
 
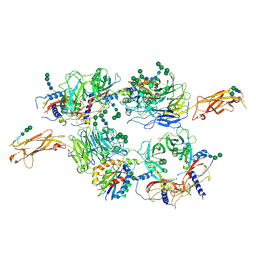 | | Integrin alpha V beta 6 in complex with pro-TGF-beta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dong, X, Zhao, B, Springer, T.A. | | Deposit date: | 2015-12-18 | | Release date: | 2017-01-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Force interacts with macromolecular structure in activation of TGF-beta.
Nature, 542, 2017
|
|
5H13
 
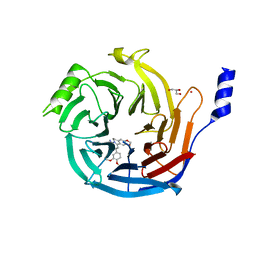 | | EED in complex with PRC2 allosteric inhibitor EED396 | | Descriptor: | 4-azanylidene-2-(3-methoxy-4-propan-2-yloxy-phenyl)-6,7-dihydro-[1,3]benzodioxolo[6,5-a]quinolizine-3-carbonitrile, GLYCEROL, PRASEODYMIUM ION, ... | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Molecular Basis of a Diverse Set of Polycomb Repressive Complex 2 Inhibitors Recognition by EED
PLoS ONE, 12, 2017
|
|
5H17
 
 | | EED in complex with PRC2 allosteric inhibitor EED210 | | Descriptor: | (3R,4aS,10aS)-6-methoxy-3-[(3-methoxyphenyl)methyl]-1-methyl-3,4,4a,5,10,10a-hexahydro-2H-benzo[g]quinoline, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and Molecular Basis of a Diverse Set of Polycomb Repressive Complex 2 Inhibitors Recognition by EED
PLoS ONE, 12, 2017
|
|
7WMW
 
 | |
7WMX
 
 | |
7WMY
 
 | |
7WMZ
 
 | |
