4BBQ
 
 | | Crystal structure of the CXXC and PHD domain of Human Lysine-specific Demethylase 2A (KDM2A)(FBXL11) | | Descriptor: | 1,2-ETHANEDIOL, LYSINE-SPECIFIC DEMETHYLASE 2A, ZINC ION | | Authors: | Allerston, C.K, Watson, A.A, Edlich, C, Li, B, Chen, Y, Ball, L, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Laue, E.D, Gileadi, O. | | Deposit date: | 2012-09-27 | | Release date: | 2012-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of the Cxxc and Phd Domain of Human Lysine-Specific Demethylase 2A (Kdm2A)(Fbxl11)
To be Published
|
|
6IDE
 
 | | Crystal structure of the Vibrio cholera VqmA-Ligand-DNA complex provides molecular mechanisms for drug design | | Descriptor: | 3,5-dimethylpyrazin-2-ol, DNA (5'-D(*AP*GP*GP*GP*GP*GP*GP*AP*AP*AP*TP*CP*CP*CP*CP*CP*CP*T)-3'), DNA (5'-D(*AP*GP*GP*GP*GP*GP*GP*AP*TP*TP*TP*CP*CP*CP*CP*CP*CP*T)-3'), ... | | Authors: | Wu, H, Li, M.J, Guo, H.J, Zhou, H, Li, B, Xu, Q, Xu, C.Y, Yu, F, He, J.H. | | Deposit date: | 2018-09-09 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of theVibrio choleraeVqmA-ligand-DNA complex provides insight into ligand-binding mechanisms relevant for drug design.
J. Biol. Chem., 294, 2019
|
|
7DAC
 
 | | Human RIPK3 amyloid fibril revealed by solid-state NMR | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Wu, X.L, Zhang, J, Dong, X.Q, Liu, J, Li, B, Hu, H, Wang, J, Wang, H.Y, Lu, J.X. | | Deposit date: | 2020-10-16 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | The structure of a minimum amyloid fibril core formed by necroptosis-mediating RHIM of human RIPK3.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8SPP
 
 | |
3KUZ
 
 | | Crystal structure of the ubiquitin like domain of PLXNC1 | | Descriptor: | Plexin-C1, UNKNOWN ATOM OR ION | | Authors: | Wang, H, Li, B, Tempel, W, Tong, Y, Guan, X, Zhong, N, Crombet, L, MacKenzie, F, Buck, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-28 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the ubiquitin like domain of PLXNC1
to be published
|
|
8W1Q
 
 | | Aerobic crystal structure of iron-bound FlcD from Pseudomonas aeruginosa | | Descriptor: | FE (II) ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Walker, M.E, Grove, T.L, Li, B, Redinbo, M.R. | | Deposit date: | 2024-02-17 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural Basis for Methine Excision by a Heme Oxygenase-like Enzyme.
Acs Cent.Sci., 10, 2024
|
|
7F59
 
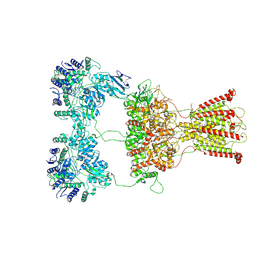 | | DNQX-bound GluK2-1xNeto2 complex | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | He, L.L, Gao, Y.W, Li, B, Zhao, Y. | | Deposit date: | 2021-06-21 | | Release date: | 2021-09-29 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Kainate receptor modulation by NETO2.
Nature, 599, 2021
|
|
7F5B
 
 | | LBD-TMD focused reconstruction of DNQX-bound GluK2-1xNeto2 complex | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | He, L.L, Gao, Y.W, Li, B, Zhao, Y. | | Deposit date: | 2021-06-21 | | Release date: | 2021-09-29 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Kainate receptor modulation by NETO2.
Nature, 599, 2021
|
|
7F56
 
 | | DNQX-bound GluK2-1xNeto2 complex, with asymmetric LBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, kainate 2, ... | | Authors: | He, L.L, Gao, Y.W, Li, B, Zhao, Y. | | Deposit date: | 2021-06-21 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Kainate receptor modulation by NETO2.
Nature, 599, 2021
|
|
7F5A
 
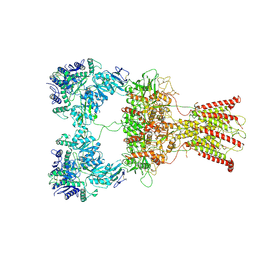 | | DNQX-bound GluK2-2xNeto2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, kainate 2, ... | | Authors: | He, L.L, Gao, Y.W, Li, B, Zhao, Y. | | Deposit date: | 2021-06-21 | | Release date: | 2021-09-29 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Kainate receptor modulation by NETO2.
Nature, 599, 2021
|
|
7F57
 
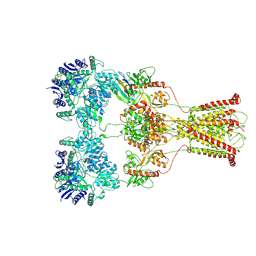 | | Kainate-bound GluK2-1xNeto2 complex, at the desensitized state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, kainate 2, ... | | Authors: | He, L.L, Gao, Y.W, Li, B, Zhao, Y. | | Deposit date: | 2021-06-21 | | Release date: | 2021-09-29 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Kainate receptor modulation by NETO2.
Nature, 599, 2021
|
|
1ZA3
 
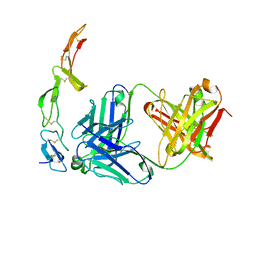 | | The crystal structure of the YSd1 Fab bound to DR5 | | Descriptor: | Fab-YSd1 heavy chain, Fab-YSd1 light chain, Tumor necrosis factor receptor superfamily member 10B | | Authors: | Fellouse, F.A, Li, B, Compaan, D.M, Peden, A.A, Hymowitz, S.G, Sidhu, S.S. | | Deposit date: | 2005-04-05 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Molecular recognition by a binary code.
J.Mol.Biol., 348, 2005
|
|
5H3J
 
 | |
5I9J
 
 | | Structure of the cholesterol and lutein-binding domain of human STARD3 at 1.74A | | Descriptor: | 1,2-ETHANEDIOL, L(+)-TARTARIC ACID, SULFATE ION, ... | | Authors: | Horvath, M.P, Bernstein, P.S, Li, B, George, E.W, Tran, Q.T. | | Deposit date: | 2016-02-20 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure of the lutein-binding domain of human StARD3 at 1.74 angstrom resolution and model of a complex with lutein.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
3SFV
 
 | | Crystal structure of the GDP-bound Rab1a S25N mutant in complex with the coiled-coil domain of LidA from Legionella pneumophila | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, LidA protein, substrate of the Dot/Icm system, ... | | Authors: | Yin, K, Lu, D, Zhu, D, Zhang, H, Li, B, Xu, S, Gu, L. | | Deposit date: | 2011-06-14 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural Insights into a Unique Legionella pneumophila Effector LidA Recognizing Both GDP and GTP Bound Rab1 in Their Active State
Plos Pathog., 8, 2012
|
|
3TKL
 
 | | Crystal structure of the GTP-bound Rab1a in complex with the coiled-coil domain of LidA from Legionella pneumophila | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, LidA protein, substrate of the Dot/Icm system, ... | | Authors: | Cheng, W, Yin, K, Lu, D, Li, B, Zhu, D, Chen, Y, Zhang, H, Xu, S, Chai, J, Gu, L. | | Deposit date: | 2011-08-27 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.183 Å) | | Cite: | Structural insights into a unique Legionella pneumophila effector LidA recognizing both GDP and GTP bound Rab1 in their active state
Plos Pathog., 8, 2012
|
|
4PT6
 
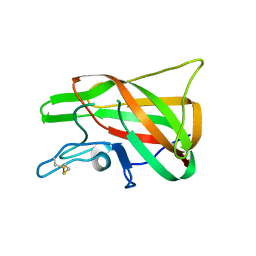 | | The discobody: an engineered discoidin domain from factor VIII that binds v 3 integrin with antibody-like affinities | | Descriptor: | Coagulation factor VIII | | Authors: | Kym, G, Shi, K, Kamajaya, A, Hamdouche, S, Kostecki, J.S, Wannier, T, Li, B, Nikolovski, P, Mayo, S.L. | | Deposit date: | 2014-03-10 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The discobody: an engineered discoidin domain from factor VIII that binds v 3 integrin with antibody-like affinities
To be Published
|
|
4LMQ
 
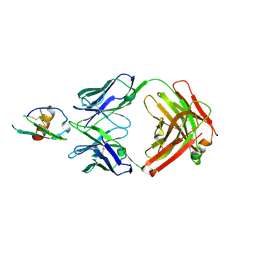 | | Development and Preclinical Characterization of a Humanized Antibody Targeting CXCL12 | | Descriptor: | Stromal cell-derived factor 1, hu30D8 Fab heavy chain, hu30D8 Fab light chain | | Authors: | Zhong, Z, Wang, J, Li, B, Xiang, H, Ultsch, M, Coons, M, Wong, T, Chiang, N.Y, Clark, S, Clark, R, Quintana, L, Gribling, P, Suto, E, Barck, K, Corpuz, R, Yao, J, Takkar, R, Lee, W.P, Damico-Beyer, L.A, Carano, R.D, Adams, C, Kelley, R.F, Wang, W, Ferrara, N. | | Deposit date: | 2013-07-10 | | Release date: | 2013-08-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.773 Å) | | Cite: | Development and Preclinical Characterization of a Humanized Antibody Targeting CXCL12.
Clin.Cancer Res., 19, 2013
|
|
6LK0
 
 | | Crystal structure of human wild type TRIP13 | | Descriptor: | Pachytene checkpoint protein 2 homolog | | Authors: | Wang, Y, Huang, J, Li, B, Xue, H, Tricot, G, Hu, L, Xu, Z, Sun, X, Chang, S, Gao, L, Tao, Y, Xu, H, Xie, Y, Xiao, W, Yu, D, Kong, Y, Chen, G, Sun, X, Lian, F, Zhang, N, Wu, X, Mao, Z, Zhan, F, Zhu, W, Shi, J. | | Deposit date: | 2019-12-17 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Small-Molecule Inhibitor Targeting TRIP13 Suppresses Multiple Myeloma Progression.
Cancer Res., 80, 2020
|
|
7W72
 
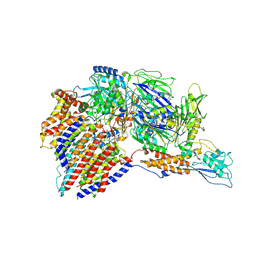 | | Structure of a human glycosylphosphatidylinositol (GPI) transamidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GPI transamidase component PIG-S, ... | | Authors: | Zhang, H, Su, J, Li, B, Gao, Y, Zhang, X.C, Zhao, Y. | | Deposit date: | 2021-12-02 | | Release date: | 2022-02-16 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of human glycosylphosphatidylinositol transamidase.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7DSV
 
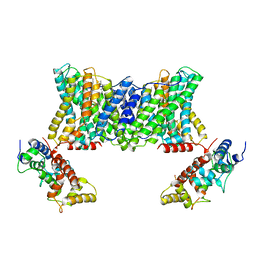 | | Structure of a human NHE1-CHP1 complex under pH 6.5 | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Calcineurin B homologous protein 1, Sodium/hydrogen exchanger 1 | | Authors: | Dong, Y, Gao, Y, Li, B, Zhang, X.C, Zhao, Y. | | Deposit date: | 2021-01-03 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and mechanism of the human NHE1-CHP1 complex.
Nat Commun, 12, 2021
|
|
7DSW
 
 | | Structure of a human NHE1-CHP1 complex under pH 7.5 | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Sodium/hydrogen exchanger 1 | | Authors: | Dong, Y, Gao, Y, Li, B, Zhang, X.C, Zhao, Y. | | Deposit date: | 2021-01-03 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and mechanism of the human NHE1-CHP1 complex.
Nat Commun, 12, 2021
|
|
7DSX
 
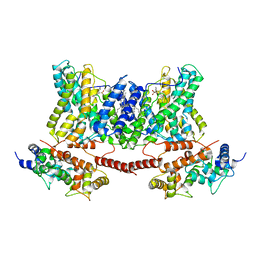 | | Structure of a human NHE1-CHP1 complex under pH 7.5, bound by cariporide | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, Calcineurin B homologous protein 1, N-[bis(azanyl)methylidene]-3-methylsulfonyl-4-propan-2-yl-benzamide, ... | | Authors: | Dong, Y, Gao, Y, Li, B, Zhang, X.C, Zhao, Y. | | Deposit date: | 2021-01-03 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and mechanism of the human NHE1-CHP1 complex.
Nat Commun, 12, 2021
|
|
7D7E
 
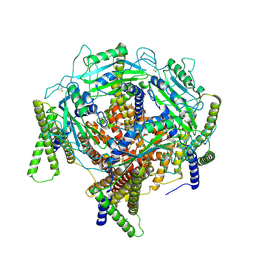 | | Structure of PKD1L3-CTD/PKD2L1 in apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Polycystic kidney disease 2-like 1 protein, ... | | Authors: | Su, Q, Chen, M, Li, B, Wang, Y, Jing, D, Zhan, X, Yu, Y, Shi, Y. | | Deposit date: | 2020-10-03 | | Release date: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for Ca 2+ activation of the heteromeric PKD1L3/PKD2L1 channel.
Nat Commun, 12, 2021
|
|
5U8T
 
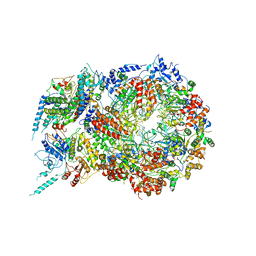 | | Structure of Eukaryotic CMG Helicase at a Replication Fork and Implications | | Descriptor: | Cell division control protein 45, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA replication complex GINS protein PSF1, ... | | Authors: | Li, B, Georgescu, R, Yuan, Z, Santos, R, Sun, J, Zhang, D, Yurieva, O, Li, H, O'Donnell, M.E. | | Deposit date: | 2016-12-15 | | Release date: | 2017-02-08 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
