2LQH
 
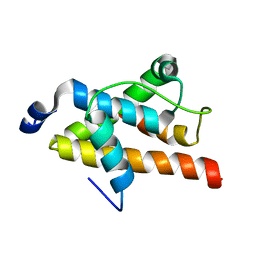 | | NMR structure of FOXO3a transactivation domains (CR2C-CR3) in complex with CBP KIX domain (2b3l conformation) | | Descriptor: | CREB-binding protein, Forkhead box O3 | | Authors: | Wang, F, Marshall, C.B, Yamamoto, K, Li, G.B, Gasmi-Seabrook, G.M.C, Okada, H, Mak, T.W, Ikura, M. | | Deposit date: | 2012-03-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures of KIX domain of CBP in complex with two FOXO3a transactivation domains reveal promiscuity and plasticity in coactivator recruitment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2MC2
 
 | |
2K60
 
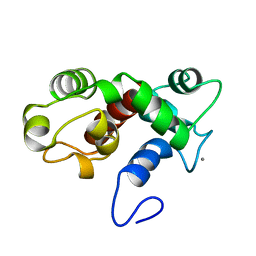 | | NMR structure of calcium-loaded STIM1 EF-SAM | | Descriptor: | CALCIUM ION, PROTEIN (Stromal interaction molecule 1) | | Authors: | Stathopulos, P.B, Ikura, M. | | Deposit date: | 2008-07-02 | | Release date: | 2008-10-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and mechanistic insights into STIM1-mediated initiation of store-operated calcium entry.
Cell(Cambridge,Mass.), 135, 2008
|
|
2L5Y
 
 | |
2KWF
 
 | | The structure of E-protein activation domain 1 bound to the KIX domain of CBP/p300 elucidates leukemia induction by E2A-PBX1 | | Descriptor: | CREB-binding protein, Transcription factor 4 | | Authors: | Denis, C.M, Chitayat, S, Plevin, M.J, Liu, S, Spencer, H.L, Ikura, M, LeBrun, D.P, Smith, S.P. | | Deposit date: | 2010-04-08 | | Release date: | 2011-07-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The structure of E-protein activation domain 1 bound to the KIX domain of CBP/p300 elucidates leukemia induction by E2A-PBX1
To be Published
|
|
5Y0D
 
 | | Crystal Structure of the human nucleosome containing the H2B E76K mutant | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Kurumizaka, H, Arimura, Y, Fujita, R, Noda, M. | | Deposit date: | 2017-07-16 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Cancer-associated mutations of histones H2B, H3.1 and H2A.Z.1 affect the structure and stability of the nucleosome.
Nucleic Acids Res., 46, 2018
|
|
5Z30
 
 | | The crystal structure of the nucleosome containing a cancer-associated histone H2A.Z R80C mutant | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A.Z, ... | | Authors: | Horikoshi, N, Arimura, Y, Kurumizaka, H. | | Deposit date: | 2018-01-05 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Cancer-associated mutations of histones H2B, H3.1 and H2A.Z.1 affect the structure and stability of the nucleosome.
Nucleic Acids Res., 46, 2018
|
|
5Y0C
 
 | | Crystal Structure of the human nucleosome at 2.09 angstrom resolution | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Kurumizaka, H, Arimura, Y, Fujita, R, Noda, M. | | Deposit date: | 2017-07-16 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.087 Å) | | Cite: | Cancer-associated mutations of histones H2B, H3.1 and H2A.Z.1 affect the structure and stability of the nucleosome.
Nucleic Acids Res., 46, 2018
|
|
5E95
 
 | | Crystal Structure of Mb(NS1)/H-Ras Complex | | Descriptor: | GTPase HRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eguchi, R.R, Sha, F, Gupta, A, Koide, A, Koide, S. | | Deposit date: | 2015-10-14 | | Release date: | 2016-11-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.402 Å) | | Cite: | Inhibition of RAS function through targeting an allosteric regulatory site.
Nat. Chem. Biol., 13, 2017
|
|
3RQR
 
 | | Crystal structure of the RYR domain of the rabbit ryanodine receptor | | Descriptor: | (UNK)(UNK)(UNK)(UNK), Ryanodine receptor 1 | | Authors: | Nair, U.B, Li, W, Dong, A, Walker, J.R, Gramolini, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-04-28 | | Release date: | 2011-06-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural determination of the phosphorylation domain of the ryanodine receptor.
Febs J., 279, 2012
|
|
7JIF
 
 | | HRAS A59T GppNHp | | Descriptor: | GLYCEROL, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Johnson, C.W, Haigis, K.M. | | Deposit date: | 2020-07-23 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.757 Å) | | Cite: | Regulation of GTPase function by autophosphorylation.
Mol.Cell, 82, 2022
|
|
7JIH
 
 | | HRAS A59E GppNHp | | Descriptor: | GLYCEROL, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Johnson, C.W, Haigis, K.M. | | Deposit date: | 2020-07-23 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | Regulation of GTPase function by autophosphorylation.
Mol.Cell, 82, 2022
|
|
7JIG
 
 | | HRAS A59T GppNHp crystal 2 | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Johnson, C.W, Haigis, K.M. | | Deposit date: | 2020-07-23 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.322 Å) | | Cite: | Regulation of GTPase function by autophosphorylation.
Mol.Cell, 82, 2022
|
|
7JII
 
 | | HRAS A59E GDP | | Descriptor: | CALCIUM ION, GTPase HRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Johnson, C.W, Haigis, K.M. | | Deposit date: | 2020-07-23 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | Regulation of GTPase function by autophosphorylation.
Mol.Cell, 82, 2022
|
|
7KMR
 
 | |
4KEI
 
 | |
4KEK
 
 | |
4KEJ
 
 | |
2Z1O
 
 | | Crystal structure of a photoswitchable GFP-like protein Dronpa in the bright-state | | Descriptor: | Fluorescent protein Dronpa | | Authors: | Kikuchi, A, Jeyakanthan, J, Taka, J, Shiro, Y, Mizuno, H, Miyawaki, A. | | Deposit date: | 2007-05-10 | | Release date: | 2008-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Light-dependent regulation of structural flexibility in a photochromic fluorescent protein
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2Z6Z
 
 | | Crystal structure of a photoswitchable GFP-like protein Dronpa in the bright-state | | Descriptor: | Fluorescent protein Dronpa | | Authors: | Kikuchi, A, Jeyakanthan, J, Taka, J, Shiro, Y, Mizuno, H, Miyawaki, A. | | Deposit date: | 2007-08-09 | | Release date: | 2008-07-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Light-dependent regulation of structural flexibility in a photochromic fluorescent protein.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2Z6X
 
 | | Crystal structure of 22G, the wild-type protein of the photoswitchable GFP-like protein Dronpa | | Descriptor: | photochromic protein Dronpa | | Authors: | Kikuchi, A, Jeyakanthan, J, Taka, J, Shiro, Y, Mizuno, H, Miyawaki, A. | | Deposit date: | 2007-08-09 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Light-dependent regulation of structural flexibility in a photochromic fluorescent protein.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2Z6Y
 
 | | Crystal structure of a photoswitchable GFP-like protein Dronpa in the bright-state | | Descriptor: | Fluorescent protein Dronpa | | Authors: | Kikuchi, A, Jeyakanthan, J, Taka, J, Shiro, Y, Mizuno, H, Miyawaki, A. | | Deposit date: | 2007-08-09 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Light-dependent regulation of structural flexibility in a photochromic fluorescent protein.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1C9B
 
 | |
1NPS
 
 | | CRYSTAL STRUCTURE OF N-TERMINAL DOMAIN OF PROTEIN S | | Descriptor: | CALCIUM ION, DEVELOPMENT-SPECIFIC PROTEIN S | | Authors: | Wenk, M, Baumgartner, R, Mayer, E.M, Huber, R, Holak, T.A, Jaenicke, R. | | Deposit date: | 1999-02-01 | | Release date: | 2000-02-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The domains of protein S from Myxococcus xanthus: structure, stability and interactions.
J.Mol.Biol., 286, 1999
|
|
8CZ7
 
 | | Crystal structure of SARS-CoV-2 Mpro with compound C2 | | Descriptor: | 3C-like proteinase, N-[(4-chlorothiophen-2-yl)methyl]-2-(isoquinolin-4-yl)-N-(4-methoxyphenyl)acetamide | | Authors: | Worrall, L.J, Lee, J, Strynadka, N.C.J. | | Deposit date: | 2022-05-24 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
