8S41
 
 | | The structure of the copia retrotransposon icosahedral capsid (T=9) | | Descriptor: | Copia VLP protein | | Authors: | Klumpe, S, Beck, F, Briggs, J.A.G, Beck, M, Plitzko, J.M. | | Deposit date: | 2024-02-20 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | In-cell structure and snapshots of copia retrotransposons in intact tissue by cryo-ET.
Cell, 188, 2025
|
|
1U7F
 
 | | Crystal Structure of the phosphorylated Smad3/Smad4 heterotrimeric complex | | Descriptor: | Mothers against decapentaplegic homolog 3, Mothers against decapentaplegic homolog 4 | | Authors: | Chacko, B.M, Qin, B.Y, Tiwari, A, Shi, G, Lam, S, Hayward, L.J, de Caestecker, M, Lin, K. | | Deposit date: | 2004-08-03 | | Release date: | 2004-09-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of heteromeric smad protein assembly in tgf-Beta signaling
Mol.Cell, 15, 2004
|
|
4YDZ
 
 | | Stress-induced protein 1 from Caenorhabditis elegans | | Descriptor: | Stress-induced protein 1 | | Authors: | Fleckenstein, T, Kastenmueller, A, Stein, M.L, Peters, C, Daake, M, Krause, M, Weinfurtner, D, Haslbeck, M, Weinkauf, S, Groll, M, Buchner, J. | | Deposit date: | 2015-02-23 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The Chaperone Activity of the Developmental Small Heat Shock Protein Sip1 Is Regulated by pH-Dependent Conformational Changes.
Mol.Cell, 58, 2015
|
|
5JDP
 
 | | E73V mutant of the human voltage-dependent anion channel | | Descriptor: | Voltage-dependent anion-selective channel protein 1 | | Authors: | Jaremko, M, Jaremko, L, Villinger, S, Schmidt, C, Giller, K, Griesinger, C, Becker, S, Zweckstetter, M. | | Deposit date: | 2016-04-17 | | Release date: | 2016-08-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | High-Resolution NMR Determination of the Dynamic Structure of Membrane Proteins.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
4YE0
 
 | | Stress-induced protein 1 truncation mutant (43 - 140) from Caenorhabditis elegans | | Descriptor: | SULFATE ION, Stress-induced protein 1 | | Authors: | Fleckenstein, T, Kastenmueller, A, Stein, M.L, Peters, C, Daake, M, Krause, M, Weinfurtner, D, Haslbeck, M, Weinkauf, S, Groll, M, Buchner, J. | | Deposit date: | 2015-02-23 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Chaperone Activity of the Developmental Small Heat Shock Protein Sip1 Is Regulated by pH-Dependent Conformational Changes.
Mol.Cell, 58, 2015
|
|
8A6M
 
 | |
7OAN
 
 | | Nanobody C5 bound to Spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Naismith, J.H, Weckener, M. | | Deposit date: | 2021-04-19 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
4FBM
 
 | | LipS and LipT, two metagenome-derived lipolytic enzymes increase the diversity of known lipase and esterase families | | Descriptor: | BROMIDE ION, LipS lipolytic enzyme | | Authors: | Chow, J, Krauss, U, Dall Antonia, Y, Fersini, F, Schmeisser, C, Schmidt, M, Menyes, I, Bornscheuer, U, Lauinger, B, Bongen, P, Pietruszka, J, Eckstein, M, Thum, O, Liese, A, Mueller-Dieckmann, J, Jaeger, K.-E, Kovavic, F, Streit, W.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Metagenome-Derived Enzymes LipS and LipT Increase the Diversity of Known Lipases.
Plos One, 7, 2012
|
|
4FBL
 
 | | LipS and LipT, two metagenome-derived lipolytic enzymes increase the diversity of known lipase and esterase families | | Descriptor: | CHLORIDE ION, LipS lipolytic enzyme, SPERMIDINE | | Authors: | Chow, J, Krauss, U, Dall Antonia, Y, Fersini, F, Schmeisser, C, Schmidt, M, Menyes, I, Bornscheuer, U, Lauinger, B, Bongen, P, Pietruszka, J, Eckstein, M, Thum, O, Liese, A, Mueller-Dieckmann, J, Jaeger, K.-E, Kovacic, F, Streit, W.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Metagenome-Derived Enzymes LipS and LipT Increase the Diversity of Known Lipases.
Plos One, 7, 2012
|
|
8BGU
 
 | | human MDM2-5S RNP | | Descriptor: | 5S rRNA, 60S ribosomal protein L11, 60S ribosomal protein L5, ... | | Authors: | Castillo, N, Thoms, M, Flemming, D, Hammaren, H.M, Buschauer, R, Ameismeier, M, Bassler, J, Beck, M, Beckmann, R, Hurt, E. | | Deposit date: | 2022-10-28 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of nascent 5S RNPs at the crossroad between ribosome assembly and MDM2-p53 pathways.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2CA7
 
 | | Conkunitzin-S1 Is The First Member Of A New Kunitz-Type Neurotoxin Family- Structural and Functional Characterization | | Descriptor: | CONKUNITZIN-S1 | | Authors: | Bayrhuber, M, Vijayan, V, Ferber, M, Graf, R, Korukottu, J, Imperial, J, Garrett, J.E, Olivera, B.M, Terlau, H, Zweckstetter, M, Becker, S. | | Deposit date: | 2005-12-19 | | Release date: | 2006-01-05 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Conkunitzin-S1 is the first member of a new Kunitz-type neurotoxin family. Structural and functional characterization.
J. Biol. Chem., 280, 2005
|
|
2JK4
 
 | | Structure of the human voltage-dependent anion channel | | Descriptor: | VOLTAGE-DEPENDENT ANION-SELECTIVE CHANNEL PROTEIN 1 | | Authors: | Bayrhuber, M, Meins, T, Habeck, M, Becker, S, Giller, K, Villinger, S, Vonrhein, C, Griesinger, C, Zweckstetter, M, Zeth, K. | | Deposit date: | 2008-08-15 | | Release date: | 2008-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structure of the Human Voltage-Dependent Anion Channel.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
6YEG
 
 | | Hybrid structure of the SPP1 tail tube by solid-state NMR and cryo EM - Final EM Refinement | | Descriptor: | Tail tube protein gp17.1* | | Authors: | Zinke, M, Sachowsky, K.A.A, Zinn-Justin, S, Ravelli, R, Schroder, G.F, Habeck, M, Lange, A. | | Deposit date: | 2020-03-24 | | Release date: | 2020-10-14 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (4 Å), SOLID-STATE NMR | | Cite: | Architecture of the flexible tail tube of bacteriophage SPP1.
Nat Commun, 11, 2020
|
|
5JAI
 
 | | Yersinia pestis FabV variant T276G | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Pschibul, A, Kuper, J, HIrschbeck, M, Kisker, C. | | Deposit date: | 2016-04-12 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selectivity of Pyridone- and Diphenyl Ether-Based Inhibitors for the Yersinia pestis FabV Enoyl-ACP Reductase.
Biochemistry, 55, 2016
|
|
5JAM
 
 | | Yersinia pestis FabV variant T276V | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DIMETHYL SULFOXIDE, Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Pschibul, A, Kuper, J, HIrschbeck, M, Kisker, C. | | Deposit date: | 2016-04-12 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selectivity of Pyridone- and Diphenyl Ether-Based Inhibitors for the Yersinia pestis FabV Enoyl-ACP Reductase.
Biochemistry, 55, 2016
|
|
5JAQ
 
 | | Yersinia pestis FabV variant T276C | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DIMETHYL SULFOXIDE, Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Pschibul, A, Kuper, J, HIrschbeck, M, Kisker, C. | | Deposit date: | 2016-04-12 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selectivity of Pyridone- and Diphenyl Ether-Based Inhibitors for the Yersinia pestis FabV Enoyl-ACP Reductase.
Biochemistry, 55, 2016
|
|
2UVS
 
 | | High Resolution Solid-state NMR structure of Kaliotoxin | | Descriptor: | POTASSIUM CHANNEL TOXIN ALPHA-KTX 3.1 | | Authors: | Korukottu, J, Lange, A, Vijayan, V, Schneider, R, Pongs, O, Becker, S, Baldus, M, Zweckstetter, M. | | Deposit date: | 2007-03-14 | | Release date: | 2008-05-27 | | Last modified: | 2024-11-06 | | Method: | SOLID-STATE NMR | | Cite: | Conformational Plasticity in Ion Channel Recognition of a Peptide Toxin
To be Published
|
|
7ABT
 
 | |
7OA6
 
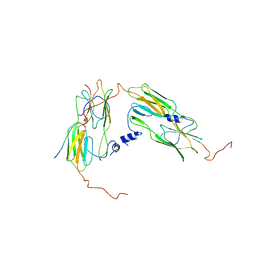 | | Pseudo-atomic model for Hsp26 residues 63 to 214. Please be advised that the target map is not of sufficient resolution to unambiguously position backbone or side chain atoms. This model represents a likely fit. | | Descriptor: | Heat shock protein 26 | | Authors: | Muehlhofer, M, Peters, C, Kriehuber, T, Kreuzeder, M, Kazman, P, Rodina, N, Reif, B, Haslbeck, M, Weinkauf, S, Buchner, J. | | Deposit date: | 2021-04-19 | | Release date: | 2021-11-24 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Phosphorylation activates the yeast small heat shock protein Hsp26 by weakening domain contacts in the oligomer ensemble.
Nat Commun, 12, 2021
|
|
6I42
 
 | |
5G2O
 
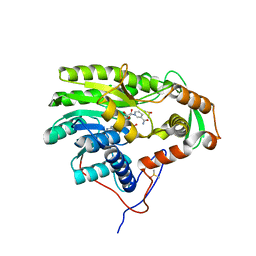 | | Yersinia pestis FabV variant T276A | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DIMETHYL SULFOXIDE, Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Pschibul, A, Kuper, J, HIrschbeck, M, Kisker, C. | | Deposit date: | 2016-04-11 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selectivity of Pyridone- and Diphenyl Ether-Based Inhibitors for the Yersinia Pestis Fabv Enoyl-Acp Reductase.
Biochemistry, 55, 2016
|
|
6P0X
 
 | |
7NY7
 
 | | Crystal structure of the Capsaspora owczarzaki macroH2A macrodomain in complex with ADP-ribose | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, Histone macroH2A1.1 | | Authors: | Guberovic, I, Knobloch, G, Basquin, J, Buschbeck, M, Ladurner, A.G. | | Deposit date: | 2021-03-21 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evolution of a histone variant involved in compartmental regulation of NAD metabolism.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7NY6
 
 | | Crystal structure of the Capsaspora owczarzaki macroH2A macrodomain | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Histone macroH2A1.1 | | Authors: | Knobloch, G, Guberovic, I, Basquin, J, Buschbeck, M, Ladurner, A.G. | | Deposit date: | 2021-03-21 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Evolution of a histone variant involved in compartmental regulation of NAD metabolism.
Nat.Struct.Mol.Biol., 28, 2021
|
|
2J80
 
 | | Structure of Citrate-bound Periplasmic Domain of Sensor Histidine Kinase CitA | | Descriptor: | CITRATE ANION, GLYCEROL, SENSOR KINASE CITA, ... | | Authors: | Sevvana, M, Vijayan, V, Zweckstetter, M, Reinelt, S, Madden, D.R, Sheldrick, G.M, Bott, M, Griesinger, C, Becker, S. | | Deposit date: | 2006-10-18 | | Release date: | 2007-10-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Ligand-Induced Switch in the Periplasmic Domain of Sensor Histidine Kinase Cita.
J.Mol.Biol., 377, 2008
|
|
