1PV6
 
 | | Crystal structure of lactose permease | | Descriptor: | Lactose permease | | Authors: | Abramson, J, Smirnova, I, Kasho, V, Verner, G, Kaback, H.R, Iwata, S. | | Deposit date: | 2003-06-26 | | Release date: | 2003-08-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and mechanism of the lactose permease of Escherichia coli
SCIENCE, 301, 2003
|
|
1PV7
 
 | | Crystal structure of lactose permease with TDG | | Descriptor: | Lactose permease, beta-D-galactopyranose-(1-1)-1-thio-beta-D-galactopyranose | | Authors: | Abramson, J, Smirnova, I, Kasho, V, Verner, G, Kaback, H.R, Iwata, S. | | Deposit date: | 2003-06-26 | | Release date: | 2003-08-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure and mechanism of the lactose permease of Escherichia coli
SCIENCE, 301, 2003
|
|
1S5L
 
 | | Architecture of the photosynthetic oxygen evolving center | | Descriptor: | 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, BETA-CAROTENE, BICARBONATE ION, ... | | Authors: | Ferreira, K.N, Iverson, T.M, Maghlaoui, K, Barber, J, Iwata, S. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-24 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Architecture of the Photosynthetic Oxygen-Evolving Center
Science, 303, 2004
|
|
6JZI
 
 | | Structure of hen egg-white lysozyme obtained from SFX experiments under atmospheric pressure | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Nango, E, Sugahara, M, Nakane, T, Tanaka, T, Iwata, S. | | Deposit date: | 2019-05-02 | | Release date: | 2019-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-viscosity sample-injection device for serial femtosecond crystallography at atmospheric pressure.
J.Appl.Crystallogr., 52, 2019
|
|
6JOD
 
 | | Angiotensin II type 2 receptor with ligand | | Descriptor: | Angiotensin II, Heavy chain of 4A03Fab, Light chain of 4A03Fab, ... | | Authors: | Asada, H, Iwata, S, Hirata, K, Shimamura, T. | | Deposit date: | 2019-03-20 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Crystal Structure of Angiotensin II Type 2 Receptor with Endogenous Peptide Hormone.
Structure, 28, 2020
|
|
6JZH
 
 | | Structure of human A2A adenosine receptor in complex with ZM241385 obtained from SFX experiments under atmospheric pressure | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Nango, E, Shimamura, T, Nakane, T, Yamanaka, Y, Mori, C, Kimura, K.T, Fujiwara, T, Tanaka, T, Iwata, S. | | Deposit date: | 2019-05-02 | | Release date: | 2019-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | High-viscosity sample-injection device for serial femtosecond crystallography at atmospheric pressure.
J.Appl.Crystallogr., 52, 2019
|
|
5B6X
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 760 ns after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-06-02 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5B6Y
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 36.2 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-06-02 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5J7A
 
 | | Bacteriorhodopsin ground state structure obtained with Serial Femtosecond Crystallography | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Nogly, P, Panneels, V, Nelson, G, Gati, C, Kimura, T, Milne, C, Milathianaki, D, Kubo, M, Wu, W, Conrad, C, Coe, J, Bean, R, Zhao, Y, Bath, P, Dods, R, Harimoorthy, R, Beyerlein, K.R, Rheinberger, J, James, D, DePonte, D, Li, C, Sala, L, Williams, G, Hunter, M, Koglin, J.E, Berntsen, P, Nango, E, Iwata, S, Chapman, H.N, Fromme, P, Frank, M, Abela, R, Boutet, S, Barty, A, White, T.A, Weierstall, U, Spence, J, Neutze, R, Schertler, G, Standfuss, J. | | Deposit date: | 2016-04-06 | | Release date: | 2016-08-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lipidic cubic phase injector is a viable crystal delivery system for time-resolved serial crystallography.
Nat Commun, 7, 2016
|
|
5B6W
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 16 ns after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-06-02 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5B6Z
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 1.725 ms us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-06-02 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5B6V
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: resting state structure | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nango, E, Royant, A, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-06-02 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
3WXQ
 
 | | Serial femtosecond X-ray structure of human fatty acid-binding protein type-3 (FABP3) in complex with stearic acid (C18:0) determined using X-ray free-electron laser at SACLA | | Descriptor: | Fatty acid-binding protein, heart, STEARIC ACID | | Authors: | Mizohata, E, Suzuki, M, Kakinouchi, K, Sugiyama, S, Murata, M, Sugahara, M, Nango, E, Tanaka, T, Tanaka, R, Tono, K, Song, C, Hatsui, T, Joti, Y, Yabashi, M, Iwata, S. | | Deposit date: | 2014-08-04 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Grease matrix as a versatile carrier of proteins for serial crystallography
Nat. Methods, 12, 2015
|
|
4APS
 
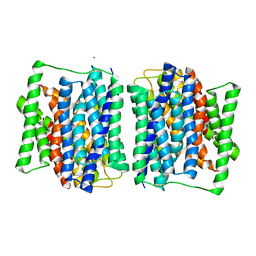 | | Crystal structure of a POT family peptide transporter in an inward open conformation. | | Descriptor: | CADMIUM ION, DI-OR TRIPEPTIDE H+ SYMPORTER | | Authors: | Solcan, N, Kwok, J, Fowler, P.W, Cameron, A.D, Drew, D, Iwata, S, Newstead, S. | | Deposit date: | 2012-04-05 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Alternating Access Mechanism in the Pot Family of Oligopeptide Transporters.
Embo J., 31, 2012
|
|
3AON
 
 | | Crystal structure of the central axis (NtpD-NtpG) in the catalytic portion of Enterococcus hirae V-type sodium ATPase | | Descriptor: | NITRATE ION, V-type sodium ATPase subunit D, V-type sodium ATPase subunit G | | Authors: | Saijo, S, Arai, S, Hossain, K.M.M, Yamato, I, Kakinuma, Y, Ishizuka-Katsura, Y, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Murata, T. | | Deposit date: | 2010-10-04 | | Release date: | 2011-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the central axis DF complex of the prokaryotic V-ATPase
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5B8C
 
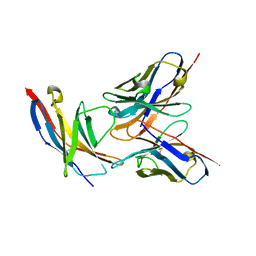 | | High resolution structure of the human PD-1 in complex with pembrolizumab Fv | | Descriptor: | Pembrolizumab heavy chain variable region (PemVH), Pembrolizumab light chain variable region (PemVL), Programmed cell death protein 1 | | Authors: | Horita, S, Shimamura, T, Iwata, S, Nomura, N. | | Deposit date: | 2016-06-14 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.146 Å) | | Cite: | High-resolution crystal structure of the therapeutic antibody pembrolizumab bound to the human PD-1
Sci Rep, 6, 2016
|
|
4CAD
 
 | | Mechanism of farnesylated CAAX protein processing by the integral membrane protease Rce1 | | Descriptor: | ANTIBODY FAB FRAGMENT HEAVY CHAIN, ANTIBODY FAB FRAGMENT LIGHT CHAIN, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Kulkarni, K, Manolaridis, I, Dodd, R.B, Cronin, N, Ogasawara, S, Iwata, S, Barford, D. | | Deposit date: | 2013-10-08 | | Release date: | 2013-11-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of Farnesylated Caax Protein Processing by the Intramembrane Protease Rce1
Nature, 504, 2013
|
|
5D4I
 
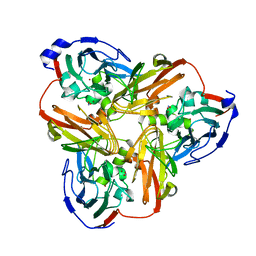 | | Intact nitrite complex of a copper nitrite reductase determined by serial femtosecond crystallography | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION | | Authors: | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Masuda, T, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-08-07 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3WXS
 
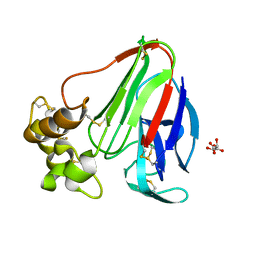 | | Thaumatin structure determined by SPring-8 Angstrom Compact free electron Laser (SACLA) | | Descriptor: | L(+)-TARTARIC ACID, thaumatin I | | Authors: | Masuda, T, Nango, E, Sugahara, M, Mizohata, E, Tanaka, T, Tanaka, R, Suzuki, M, Mikami, B, Iwata, S. | | Deposit date: | 2014-08-07 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Grease matrix as a versatile carrier of proteins for serial crystallography
Nat. Methods, 12, 2015
|
|
5NDC
 
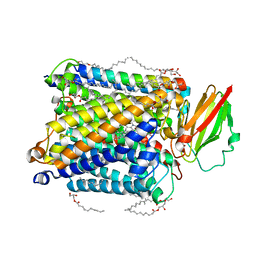 | | Structure of ba3-type cytochrome c oxidase from Thermus thermophilus by serial femtosecond crystallography | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide IIA, ... | | Authors: | Andersson, R, Safari, C, Dods, R, Nango, E, Tanaka, R, Nakane, T, Tono, K, Joti, Y, Bath, P, Dunevall, E, Bosman, E, Nureki, O, Iwata, S, Neutze, R, Branden, G. | | Deposit date: | 2017-03-08 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Serial femtosecond crystallography structure of cytochrome c oxidase at room temperature.
Sci Rep, 7, 2017
|
|
3AIE
 
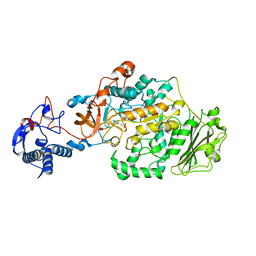 | | Crystal Structure of glucansucrase from Streptococcus mutans | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Glucosyltransferase-SI | | Authors: | Ito, K, Ito, S, Shimamura, T, Iwata, S. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans.
J.Mol.Biol., 408, 2011
|
|
2CFQ
 
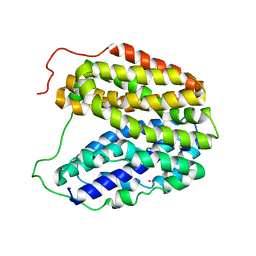 | | Sugar Free Lactose Permease at neutral pH | | Descriptor: | LACTOSE PERMEASE, MERCURY (II) ION | | Authors: | Mirza, O, Guan, L, Verner, G, Iwata, S, Kaback, H.R. | | Deposit date: | 2006-02-22 | | Release date: | 2006-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Evidence for Induced Fit and a Mechanism for Sugar/H(+) Symport in Lacy.
Embo J., 25, 2006
|
|
4ATV
 
 | | STRUCTURE OF A TRIPLE MUTANT OF THE NHAA DIMER, CRYSTALLISED AT LOW PH | | Descriptor: | DODECYL-ALPHA-D-MALTOSIDE, NA(+)/H(+) ANTIPORTER NHAA, SULFATE ION | | Authors: | Drew, D, Lee, C, Iwata, S, Cameron, A.D. | | Deposit date: | 2012-05-10 | | Release date: | 2013-07-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the sodium-proton antiporter NhaA dimer and new mechanistic insights.
J. Gen. Physiol., 144, 2014
|
|
4AU5
 
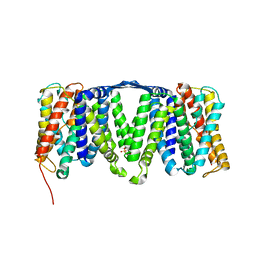 | | Structure of the NhaA dimer, crystallised at low pH | | Descriptor: | DODECYL-ALPHA-D-MALTOSIDE, NA(+)/H(+) ANTIPORTER NHAA, SULFATE ION | | Authors: | Drew, D, Lee, C, Iwata, S, Cameron, A.D. | | Deposit date: | 2012-05-14 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.696 Å) | | Cite: | Crystal structure of the sodium-proton antiporter NhaA dimer and new mechanistic insights.
J. Gen. Physiol., 144, 2014
|
|
3AIB
 
 | | Crystal Structure of Glucansucrase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Glucosyltransferase-SI, ... | | Authors: | Ito, K, Ito, S, Shimamura, T, Iwata, S. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans.
J.Mol.Biol., 408, 2011
|
|
