7NKY
 
 | | RNA Polymerase II-Spt4/5-nucleosome-FACT structure | | Descriptor: | Chromatin elongation factor SPT4, DNA (138-MER), DNA (148-MER), ... | | Authors: | Farnung, L, Ochmann, M, Engeholm, M, Cramer, P. | | Deposit date: | 2021-02-19 | | Release date: | 2021-07-07 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of nucleosome transcription mediated by Chd1 and FACT.
Nat.Struct.Mol.Biol., 28, 2021
|
|
4ZKN
 
 | |
7NN3
 
 | | A carbohydrate esterase family 15 (CE15) glucuronoyl esterase from Caldicellulosiruptor kristjansonii | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Beta-xylanase, ... | | Authors: | Krska, D, Mazurkewich, S, Navarro Poulsen, J, Larsbrink, J, Lo Leggio, L. | | Deposit date: | 2021-02-24 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88948345 Å) | | Cite: | Structural and Functional Analysis of a Multimodular Hyperthermostable Xylanase-Glucuronoyl Esterase from Caldicellulosiruptor kristjansonii .
Biochemistry, 60, 2021
|
|
4ZQC
 
 | | Tryptophan Synthase from Salmonella typhimurium in complex with two molecules of N-(4'-trifluoromethoxybenzoyl)-2-amino-1-ethylphosphate (F6F) inhibitor in the alpha-site and a single F6F molecule in the beta-site at 1.54 Angstrom resolution. | | Descriptor: | 2-{[4-(TRIFLUOROMETHOXY)BENZOYL]AMINO}ETHYL DIHYDROGEN PHOSPHATE, DIMETHYL SULFOXIDE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Hilario, E, Caulkins, B.G, Young, R.P, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2015-05-08 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Visualizing the tunnel in tryptophan synthase with crystallography: Insights into a selective filter for accommodating indole and rejecting water.
Biochim.Biophys.Acta, 1864, 2016
|
|
7N98
 
 | | Cryo-EM structure of MFSD2A | | Descriptor: | Sodium-dependent lysophosphatidylcholine symporter 1 | | Authors: | Zhang, J, Feng, L. | | Deposit date: | 2021-06-17 | | Release date: | 2021-08-04 | | Last modified: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and mechanism of blood-brain-barrier lipid transporter MFSD2A.
Nature, 596, 2021
|
|
7MWC
 
 | | ERAP1 binds peptide C-terminus of a LPF sequence (AAAAFKARKF) | | Descriptor: | Endoplasmic reticulum aminopeptidase 1,LPF sequence | | Authors: | Guo, H.C, Sui, L. | | Deposit date: | 2021-05-16 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | ERAP1 binds peptide C-termini of different sequences and/or lengths by a common recognition mechanism.
Immunobiology, 226, 2021
|
|
2I04
 
 | | X-ray crystal structure of MAGI-1 PDZ1 bound to the C-terminal peptide of HPV18 E6 | | Descriptor: | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1, SULFATE ION, ... | | Authors: | Chen, X.S, Zhang, Y, Dasgupta, J, Banks, L, Thomas, M. | | Deposit date: | 2006-08-09 | | Release date: | 2007-02-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of a Human Papillomavirus (HPV) E6 Polypeptide Bound to MAGUK Proteins: Mechanisms of Targeting Tumor Suppressors by a High-Risk HPV Oncoprotein.
J.Virol., 81, 2007
|
|
5G4H
 
 | | 1.50 A resolution catechol (1,2-dihydroxybenzene) inhibited Sporosarcina pasteurii urease | | Descriptor: | 1,2-ETHANEDIOL, CATECHOL, HYDROXIDE ION, ... | | Authors: | Mazzei, L, Cianci, M, Musiani, F, Ciurli, S. | | Deposit date: | 2016-05-13 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Inactivation of Urease by Catechol: Kinetics and Structure.
J.Inorg.Biochem., 166, 2016
|
|
2I0L
 
 | | X-ray crystal structure of Sap97 PDZ2 bound to the C-terminal peptide of HPV18 E6. | | Descriptor: | Disks large homolog 1, peptide E6 | | Authors: | Chen, X.S, Zhang, Y, Dasgupta, J, Banks, L, Thomas, M. | | Deposit date: | 2006-08-10 | | Release date: | 2007-02-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structures of a Human Papillomavirus (HPV) E6 Polypeptide Bound to MAGUK Proteins: Mechanisms of Targeting Tumor Suppressors by a High-Risk HPV Oncoprotein.
J.Virol., 81, 2007
|
|
5G4T
 
 | |
7NKX
 
 | | RNA polymerase II-Spt4/5-nucleosome-Chd1 structure | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromatin elongation factor SPT4, ... | | Authors: | Farnung, L, Ochmann, M, Engeholm, M, Cramer, P. | | Deposit date: | 2021-02-19 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of nucleosome transcription mediated by Chd1 and FACT.
Nat.Struct.Mol.Biol., 28, 2021
|
|
4ZLR
 
 | | Structure of the Brat-NHL domain bound to consensus RNA motif | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Brain tumor protein, PENTAETHYLENE GLYCOL, ... | | Authors: | Jakob, L, Treiber, N, Treiber, T, Stotz, M, Loedige, I, Meister, G. | | Deposit date: | 2015-05-01 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of the NHL Domain in Complex with RNA Reveals the Molecular Basis of Drosophila Brain-Tumor-Mediated Gene Regulation.
Cell Rep, 13, 2015
|
|
7N3O
 
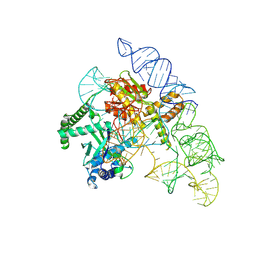 | | Cryo-EM structure of the Cas12k-sgRNA complex | | Descriptor: | Cas12k, Single guide RNA | | Authors: | Chang, L, Li, Z, Xiao, R, Wang, S, Han, R. | | Deposit date: | 2021-06-01 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of target DNA recognition by CRISPR-Cas12k for RNA-guided DNA transposition.
Mol.Cell, 81, 2021
|
|
4ZMU
 
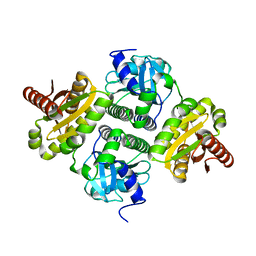 | | Dcsbis, a diguanylate cyclase from Pseudomonas aeruginosa | | Descriptor: | diguanylate cyclase | | Authors: | Chen, Y, Liu, C, Liu, S, Chi, K, Gu, L. | | Deposit date: | 2015-05-04 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | crystal structure of Dcsbis from Pseudomonas aeruginosa
To Be Published
|
|
7N3P
 
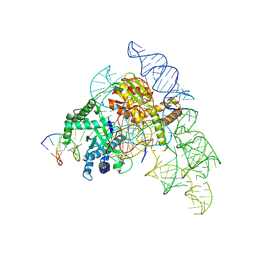 | | Cryo-EM structure of the Cas12k-sgRNA-dsDNA complex | | Descriptor: | Cas12k, DNA (5'-D(*CP*AP*TP*GP*AP*CP*TP*TP*CP*TP*CP*AP*AP*CP*CP*GP*AP*GP*TP*TP*T)-3'), DNA (5'-D(P*AP*AP*AP*CP*TP*CP*GP*GP*TP*T)-3'), ... | | Authors: | Chang, L, Li, Z, Xiao, R, Wang, S, Han, R. | | Deposit date: | 2021-06-01 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural basis of target DNA recognition by CRISPR-Cas12k for RNA-guided DNA transposition.
Mol.Cell, 81, 2021
|
|
4ZPQ
 
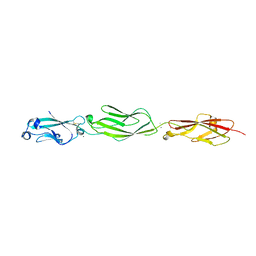 | | Crystal Structure of Protocadherin Gamma C5 EC1-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, MCG133388, ... | | Authors: | Wolcott, H.N, Goodman, K.M, Bahna, F, Mannepalli, S, Shapiro, L. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.099 Å) | | Cite: | Molecular Logic of Neuronal Self-Recognition through Protocadherin Domain Interactions.
Cell, 163, 2015
|
|
4ZQM
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the complex with XMP and NAD | | Descriptor: | Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, XANTHOSINE-5'-MONOPHOSPHATE | | Authors: | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-10 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Mycobacterium tuberculosis IMPDH in Complexes with Substrates, Products and Antitubercular Compounds.
Plos One, 10, 2015
|
|
7N7W
 
 | | Crystal Structure of SARS-CoV-2 NendoU in complex with CSC000178569 | | Descriptor: | N-(2-fluorophenyl)-N'-methylurea, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-06-11 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 51, 2023
|
|
7N83
 
 | | Crystal Structure of SARS-CoV-2 NendoU in complex with Z2443429438 | | Descriptor: | (3S)-1-(phenylsulfonyl)pyrrolidin-3-amine, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-06-12 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 51, 2023
|
|
5HGE
 
 | | Filamentous Assembly of Green Fluorescent Protein Supported by a C-terminal fusion of 18-residues, viewed in space group P212121 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Green fluorescent protein | | Authors: | Sawaya, M.R, Heller, D.M, McPartland, L, Hochschild, A, Eisenberg, D.S. | | Deposit date: | 2016-01-08 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.863 Å) | | Cite: | Green Fluorescent Protein Fusion that Self Assembles as Polar Filaments
to be published
|
|
4ZRA
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS LPRG BINDING TO TRIACYLGLYCERIDE | | Descriptor: | Lipoprotein LprG, Tripalmitoylglycerol | | Authors: | Martinot, A.J, Farrow, M, Bai, L, Layre, E, Cheng, T.Y, Tsai, J.H.C, Iqbal, J, Annand, J, Sullivan, Z, Hussain, M, Sacchettini, J, Moody, D.B, Seeliger, J, Rubin, E.J, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-05-12 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Mycobacterial Metabolic Syndrome: LprG and Rv1410 Regulate Triacylglyceride Levels, Growth Rate and Virulence in Mycobacterium tuberculosis.
Plos Pathog., 12, 2016
|
|
7N7Y
 
 | | Crystal Structure of SARS-CoV-2 NendoU in complex with Z18197050 | | Descriptor: | Uridylate-specific endoribonuclease, methyl 4-sulfamoylbenzoate | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-06-11 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 51, 2023
|
|
7N7U
 
 | | Crystal Structure of SARS-CoV-2 NendoU in complex with LIZA-7 | | Descriptor: | 1-[(2~{R},4~{S},5~{R})-5-[[(azanylidene-$l^{4}-azanylidene)amino]methyl]-4-oxidanyl-oxolan-2-yl]-5-methyl-pyrimidine-2,4-dione, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-06-11 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 51, 2023
|
|
4ZPL
 
 | | Crystal Structure of Protocadherin Beta 1 EC1-3 | | Descriptor: | CALCIUM ION, Protein Pcdhb1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Goodman, K.M, Bahna, F, Shapiro, L. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Molecular Logic of Neuronal Self-Recognition through Protocadherin Domain Interactions.
Cell, 163, 2015
|
|
5HJ1
 
 | | Crystal structure of PDZ domain of pullulanase C protein of type II secretion system from Klebsiella pneumoniae in complex with fatty acid | | Descriptor: | Pullulanase C protein, VACCENIC ACID | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-12 | | Release date: | 2016-02-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of PDZ domain of pullulanase C protein of type II secretion system from Klebsiella pneumoniae in complex with fatty acid
To Be Published
|
|
