5W9M
 
 | |
2QLH
 
 | | mPlum I65L mutant | | Descriptor: | Fluorescent protein plum | | Authors: | Shu, X, Remington, S.J. | | Deposit date: | 2007-07-12 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies of Far-Red Emission in mPlum, a Monomeric Red Fluorescent Protein
To be Published
|
|
4YES
 
 | |
7CWO
 
 | | SARS-CoV-2 spike protein RBD and P17 fab complex | | Descriptor: | Spike glycoprotein, heavy chain of P17 Fab, light chain of P17 Fab | | Authors: | Wang, X, Wang, N. | | Deposit date: | 2020-08-29 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Rational development of a human antibody cocktail that deploys multiple functions to confer Pan-SARS-CoVs protection.
Cell Res., 31, 2021
|
|
6WJ5
 
 | | Structure of human TRPA1 in complex with inhibitor GDC-0334 | | Descriptor: | (4R,5S)-4-fluoro-1-[(4-fluorophenyl)sulfonyl]-5-methyl-N-({5-(trifluoromethyl)-2-[2-(trifluoromethyl)pyrimidin-5-yl]pyridin-4-yl}methyl)-L-prolinamide, Transient receptor potential cation channel subfamily A member 1 | | Authors: | Rohou, A, Rouge, L, Arthur, C.P, Volgraf, M, Chen, H. | | Deposit date: | 2020-04-11 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A TRPA1 inhibitor suppresses neurogenic inflammation and airway contraction for asthma treatment.
J.Exp.Med., 218, 2021
|
|
7MGK
 
 | |
2QLI
 
 | | mPlum E16Q mutant | | Descriptor: | Fluorescent protein plum | | Authors: | Shu, X, Remington, S.J. | | Deposit date: | 2007-07-12 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structural Studies of Far-Red Emission in mPlum, a Monomeric Red Fluorescent Protein
To be Published
|
|
7MGJ
 
 | |
7CWT
 
 | |
8E7E
 
 | |
8E7J
 
 | |
4FWI
 
 | | Crystal structure of the nucleotide-binding domain of a dipeptide ABC transporter | | Descriptor: | ABC-type dipeptide/oligopeptide/nickel transport system, ATPase component, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Li, X, Ge, J, Yang, M, Wang, N. | | Deposit date: | 2012-07-01 | | Release date: | 2013-01-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.892 Å) | | Cite: | Structure of the nucleotide-binding domain of a dipeptide ABC transporter reveals a novel iron-sulfur cluster-binding domain
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7BQY
 
 | | THE CRYSTAL STRUCTURE OF COVID-19 MAIN PROTEASE IN COMPLEX WITH AN INHIBITOR N3 at 1.7 angstrom | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Liu, X, Zhang, B, Jin, Z, Yang, H, Rao, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors.
Nature, 582, 2020
|
|
4XWW
 
 | | Crystal structure of RNase J complexed with RNA | | Descriptor: | DR2417, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Lu, M, Zhang, H, Xu, Q, Hua, Y, Zhao, Y. | | Deposit date: | 2015-01-29 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into catalysis and dimerization enhanced exonuclease activity of RNase J
Nucleic Acids Res., 43, 2015
|
|
7W5Y
 
 | |
7W5X
 
 | | Cryo-EM structure of SoxS-dependent transcription activation complex with zwf promoter DNA | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Lin, W, Feng, Y, Shi, J. | | Deposit date: | 2021-11-30 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of three different transcription activation strategies adopted by a single regulator SoxS.
Nucleic Acids Res., 50, 2022
|
|
7W5W
 
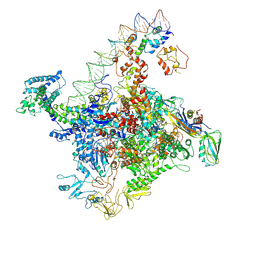 | |
4JOL
 
 | |
7CWU
 
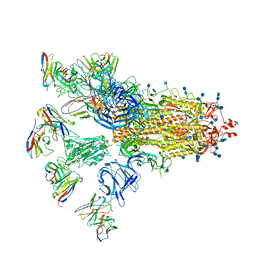 | | SARS-CoV-2 spike proteins trimer in complex with P17 and FC05 Fabs cocktail | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Wang, N. | | Deposit date: | 2020-08-31 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure-based development of human antibody cocktails against SARS-CoV-2.
Cell Res., 31, 2021
|
|
7CWN
 
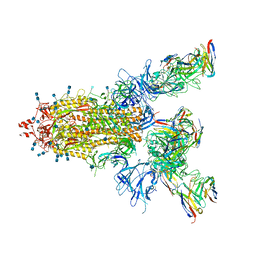 | | P17-H014 Fab cocktail in complex with SARS-CoV-2 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, N, Wang, X. | | Deposit date: | 2020-08-29 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Rational development of a human antibody cocktail that deploys multiple functions to confer Pan-SARS-CoVs protection.
Cell Res., 31, 2021
|
|
6IVE
 
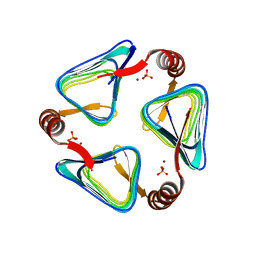 | |
7CMW
 
 | | Complex structure of PARP1 catalytic domain with pamiparib | | Descriptor: | (2R)-14-fluoro-2-methyl-6,9,10,19-tetrazapentacyclo[14.2.1.02,6.08,18.012,17]nonadeca-1(18),8,12(17),13,15-pentaen-11-one, GLYCEROL, Poly [ADP-ribose] polymerase 1 | | Authors: | Feng, Y.C, Peng, H, Hong, Y, Liu, Y. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Pamiparib (BGB-290), a Potent and Selective Poly (ADP-ribose) Polymerase (PARP) Inhibitor in Clinical Development.
J.Med.Chem., 63, 2020
|
|
7XB0
 
 | | Crystal structure of Omicron BA.2 RBD complexed with hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, L, Liao, H, Meng, Y, Li, W. | | Deposit date: | 2022-03-19 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.
Cell, 185, 2022
|
|
4KFO
 
 | | Structure-Based Discovery of Novel Amide-Containing Nicotinamide Phosphoribosyltransferase (Nampt) Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, N-{4-[(3,5-difluorophenyl)sulfonyl]benzyl}imidazo[1,2-a]pyridine-6-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Zheng, X, Bauer, P, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Han, B, Ho, Y, Kley, N, Lin, J, Reynolds, D.J, Sharma, G, Smith, C.C, Wang, Z, Dragovich, P.S, Gunzner-Tosteb, J, Liederer, B.M, Ly, J, O'Brien, T, Oh, A, Wang, L, Wang, W, Xiao, Y, Zak, M, Zhao, G, Yuen, P, Bair, K.W. | | Deposit date: | 2013-04-27 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based identification of ureas as novel nicotinamide phosphoribosyltransferase (nampt) inhibitors.
J.Med.Chem., 56, 2013
|
|
2QLG
 
 | | mPlum | | Descriptor: | Fluorescent protein plum | | Authors: | Shu, X, Remington, S.J. | | Deposit date: | 2007-07-12 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Studies of Far-Red Emission in mPlum, a Monomeric Red Fluorescent Protein
To be Published
|
|
