6U9K
 
 | | MLL1 SET N3861I/Q3867L bound to inhibitor 18 (TC-5153) | | Descriptor: | 5'-([(3S)-3-amino-3-carboxypropyl]{[1-(3,3-diphenylpropyl)azetidin-3-yl]methyl}amino)-5'-deoxyadenosine, GLYCEROL, Histone-lysine N-methyltransferase, ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Potent Small-Molecule Inhibitors of MLL Methyltransferase.
Acs Med.Chem.Lett., 11, 2020
|
|
6U9N
 
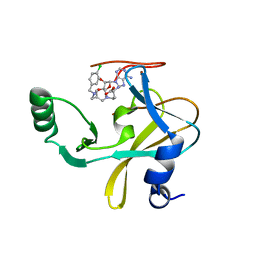 | | MLL1 SET N3861I/Q3867L bound to inhibitor 14 (TC-5139) | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl]({1-[(4-chlorophenyl)methyl]azetidin-3-yl}methyl)amino}-5'-deoxyadenosine, Histone-lysine N-methyltransferase, ZINC ION | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of Potent Small-Molecule Inhibitors of MLL Methyltransferase.
Acs Med.Chem.Lett., 11, 2020
|
|
6U9R
 
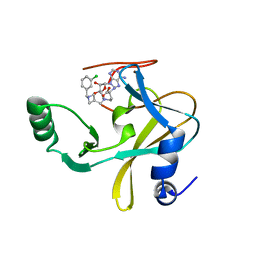 | | MLL1 SET N3861I/Q3867L bound to inhibitor 12 (TC-5140) | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl]({1-[(3-chlorophenyl)methyl]azetidin-3-yl}methyl)amino}-5'-deoxyadenosine, Histone-lysine N-methyltransferase, ZINC ION | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Potent Small-Molecule Inhibitors of MLL Methyltransferase.
Acs Med.Chem.Lett., 11, 2020
|
|
6O96
 
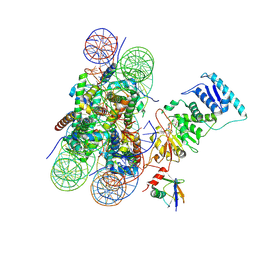 | | Dot1L bound to the H2BK120 Ubiquitinated nucleosome | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Valencia-Sanchez, M.I, De Ioannes, P.E, Miao, W, Vasilyev, N, Chen, R, Nudler, E, Armache, J.-P, Armache, K.-J. | | Deposit date: | 2019-03-13 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Basis of Dot1L Stimulation by Histone H2B Lysine 120 Ubiquitination.
Mol.Cell, 74, 2019
|
|
6U9M
 
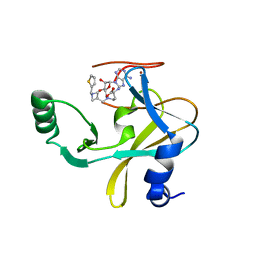 | | MLL1 SET N3861I/Q3867L bound to inhibitor 16 (TC-5109) | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl]({1-[(thiophen-2-yl)methyl]azetidin-3-yl}methyl)amino}-5'-deoxyadenosine, GLYCEROL, Histone-lysine N-methyltransferase, ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Potent Small-Molecule Inhibitors of MLL Methyltransferase.
Acs Med.Chem.Lett., 11, 2020
|
|
3S5J
 
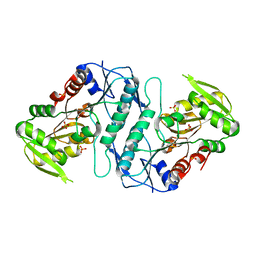 | |
7RW2
 
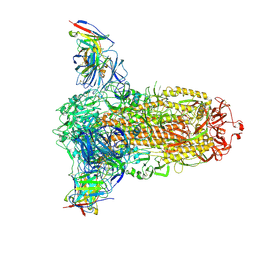 | |
3HYF
 
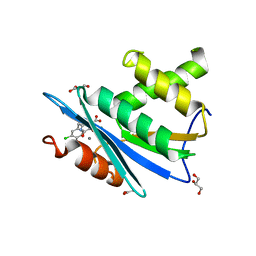 | | Crystal structure of HIV-1 RNase H p15 with engineered E. coli loop and active site inhibitor | | Descriptor: | 2-(3,4-dichlorobenzyl)-5,6-dihydroxypyrimidine-4-carboxylic acid, ACETATE ION, GLYCEROL, ... | | Authors: | Lansdon, E.B, Kirschberg, T.A. | | Deposit date: | 2009-06-22 | | Release date: | 2009-10-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | RNase H active site inhibitors of human immunodeficiency virus type 1 reverse transcriptase: design, biochemical activity, and structural information.
J.Med.Chem., 52, 2009
|
|
6T6B
 
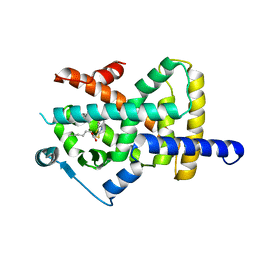 | | Crystal structure of PPARgamma in complex with compound 16 (MF27) | | Descriptor: | (2~{R})-2-[[6-[(2,4-dichlorophenyl)sulfonylamino]-1,3-benzothiazol-2-yl]sulfanyl]octanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Ni, X, Hanke, T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-18 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Selective Modulator of Peroxisome Proliferator-Activated Receptor gamma with an Unprecedented Binding Mode.
J.Med.Chem., 63, 2020
|
|
6ETZ
 
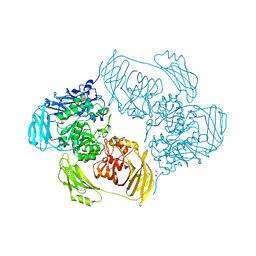 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB | | Descriptor: | ACETATE ION, Beta-galactosidase, MALONATE ION, ... | | Authors: | Rutkiewicz-Krotewicz, M, Bujacz, A, Bujacz, G. | | Deposit date: | 2017-10-27 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In Situ Random Microseeding and Streak Seeding Used for Growth of Crystals of Cold-Adapted Beta-D-Galactosidases: Crystal Structure of BetaDG from Arthrobacter sp. 32cB
Crystals, 8, 2018
|
|
5XWP
 
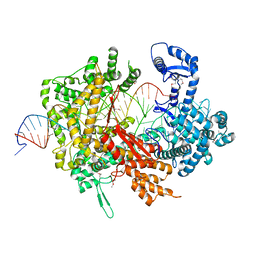 | | Crystal structure of LbuCas13a-crRNA-target RNA ternary complex | | Descriptor: | RNA (30-MER), RNA (59-MER), Uncharacterized protein | | Authors: | Liu, L, Li, X, Li, Z, Wang, Y. | | Deposit date: | 2017-06-30 | | Release date: | 2017-09-13 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3.086 Å) | | Cite: | The Molecular Architecture for RNA-Guided RNA Cleavage by Cas13a.
Cell, 170, 2017
|
|
5GNF
 
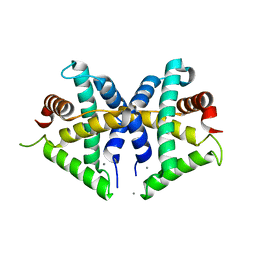 | | Crystal structure of anti-CRISPR protein AcrF3 | | Descriptor: | CALCIUM ION, Uncharacterized protein AcrF3 | | Authors: | Wang, J, Wang, Y. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A CRISPR evolutionary arms race: structural insights into viral anti-CRISPR/Cas responses
Cell Res., 26, 2016
|
|
5GQH
 
 | | Cryo-EM structure of PaeCas3-AcrF3 complex | | Descriptor: | CRISPR-associated nuclease/helicase Cas3 subtype I-F/YPEST, anti-CRISPR protein 3 | | Authors: | Zhang, X, Ma, J, Wang, Y, Wang, J. | | Deposit date: | 2016-08-07 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A CRISPR evolutionary arms race: structural insights into viral anti-CRISPR/Cas responses
Cell Res., 26, 2016
|
|
2WSO
 
 | | Structure of Cerulean Fluorescent Protein at physiological pH | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | Lelimousin, M, Noirclerc-Savoye, M, Lazareno-Saez, C, Paetzold, B, Le Vot, S, Chazal, R, Macheboeuf, P, Field, M.J, Bourgeois, D, Royant, A. | | Deposit date: | 2009-09-08 | | Release date: | 2009-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Intrinsic Dynamics in Ecfp and Cerulean Control Fluorescence Quantum Yield.
Biochemistry, 48, 2009
|
|
7BEG
 
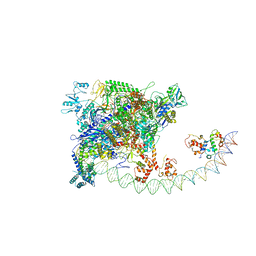 | | Structures of class I bacterial transcription complexes | | Descriptor: | Class I pacrA promoter non-template DNA, Class I pacrA promoter template DNA, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Ye, F.Z, Hao, M, Zhang, X.D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structures of Class I and Class II Transcription Complexes Reveal the Molecular Basis of RamA-Dependent Transcription Activation.
Adv Sci, 9, 2022
|
|
7BEF
 
 | | Structures of class II bacterial transcription complexes | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Hao, M, Ye, F.Z, Zhang, X.D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structures of Class I and Class II Transcription Complexes Reveal the Molecular Basis of RamA-Dependent Transcription Activation.
Adv Sci, 9, 2022
|
|
2OSH
 
 | |
6JIR
 
 | | Crystal structure of C. crescentus beta sliding clamp with PEG bound to putative beta-motif tethering region | | Descriptor: | 1,2-ETHANEDIOL, Beta sliding clamp, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Jiang, X, Teng, M, Li, X. | | Deposit date: | 2019-02-23 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Caulobacter crescentus beta sliding clamp employs a noncanonical regulatory model of DNA replication.
Febs J., 287, 2020
|
|
3LAL
 
 | | Crystal structure of HIV-1 reverse transcriptase in complex with N1-ethyl pyrimidinedione non-nucleoside inhibitor | | Descriptor: | 3-{[3-ethyl-5-(1-methylethyl)-2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl]carbonyl}-5-methylbenzonitrile, HIV Reverse transcriptase, SULFATE ION | | Authors: | Lansdon, E.B, Mitchell, M.L. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | N1-Alkyl pyrimidinediones as non-nucleoside inhibitors of HIV-1 reverse transcriptase.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3LAK
 
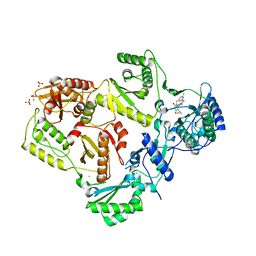 | | Crystal structure of HIV-1 reverse transcriptase in complex with N1-heterocycle pyrimidinedione non-nucleoside inhibitor | | Descriptor: | 3-({3-[(2-amino-6-fluoropyridin-4-yl)methyl]-5-(1-methylethyl)-2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl}carbonyl)-5-methylbenzonitrile, CHLORIDE ION, HIV Reverse transcriptase, ... | | Authors: | Lansdon, E.B, Mitchell, M.L. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | N1-Heterocyclic pyrimidinediones as non-nucleoside inhibitors of HIV-1 reverse transcriptase.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3LAM
 
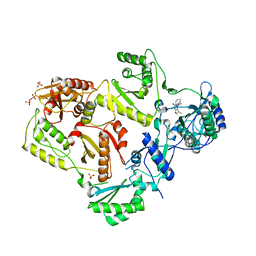 | | Crystal structure of HIV-1 reverse transcriptase in complex with N1-propyl pyrimidinedione non-nucleoside inhibitor | | Descriptor: | 3-methyl-5-{[5-(1-methylethyl)-2,6-dioxo-3-propyl-1,2,3,6-tetrahydropyrimidin-4-yl]carbonyl}benzonitrile, HIV Reverse transcriptase, SULFATE ION | | Authors: | Lansdon, E.B, Mitchell, M.L. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | N1-Alkyl pyrimidinediones as non-nucleoside inhibitors of HIV-1 reverse transcriptase.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3LAN
 
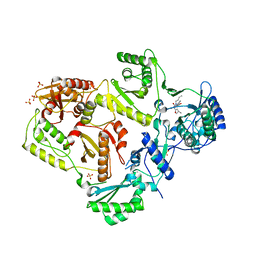 | | Crystal structure of HIV-1 reverse transcriptase in complex with N1-butyl pyrimidinedione non-nucleoside inhibitor | | Descriptor: | 3-{[3-butyl-5-(1-methylethyl)-2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl]carbonyl}-5-methylbenzonitrile, HIV Reverse transcriptase, SULFATE ION | | Authors: | Lansdon, E.B, Mitchell, M.L. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | N1-Alkyl pyrimidinediones as non-nucleoside inhibitors of HIV-1 reverse transcriptase.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7UC6
 
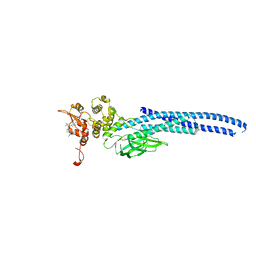 | | Stat5a Core in complex with Compound 12 | | Descriptor: | N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N~3~-(4-bromophenyl)-N,N-dimethyl-beta-alaninamide, Signal transducer and activator of transcription 5A | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2022-03-16 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Discovery of a Potent and Selective STAT5 PROTAC Degrader with Strong Antitumor Activity In Vivo in Acute Myeloid Leukemia.
J.Med.Chem., 66, 2023
|
|
7UBT
 
 | | Stat5a Core in complex with Compound 18 | | Descriptor: | GLYCEROL, N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N~3~-(1,3-benzothiazol-5-yl)-N,N-dimethyl-beta-alaninamide, Signal transducer and activator of transcription 5A | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2022-03-15 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | Discovery of a Potent and Selective STAT5 PROTAC Degrader with Strong Antitumor Activity In Vivo in Acute Myeloid Leukemia.
J.Med.Chem., 66, 2023
|
|
7UC7
 
 | | Stat5a Core in complex with Compound 17 | | Descriptor: | GLYCEROL, N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N~3~-(1-benzofuran-5-yl)-N,N-dimethyl-beta-alaninamide, Signal transducer and activator of transcription 5A | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2022-03-16 | | Release date: | 2023-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Discovery of a Potent and Selective STAT5 PROTAC Degrader with Strong Antitumor Activity In Vivo in Acute Myeloid Leukemia.
J.Med.Chem., 66, 2023
|
|
