5UFR
 
 | | Structure of RORgt bound to | | Descriptor: | (S)-[4-chloro-2-(dimethylamino)-3-phenylquinolin-6-yl](1-methyl-1H-imidazol-5-yl)(pyridin-4-yl)methanol, Nuclear receptor ROR-gamma | | Authors: | Spurlino, J, Abad, M. | | Deposit date: | 2017-01-05 | | Release date: | 2017-04-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.068 Å) | | Cite: | Identification and structure activity relationships of quinoline tertiary alcohol modulators of ROR gamma t.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
1TFB
 
 | |
3P0B
 
 | |
7DE2
 
 | | iron and alpha-ketoglutarate-dependent endoperoxidase NvfI | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, NvfI, ... | | Authors: | Zhai, R, Mori, T, Abe, I. | | Deposit date: | 2020-11-01 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular insights into the endoperoxide formation by Fe(II)/ alpha-KG-dependent oxygenase NvfI.
Nat Commun, 12, 2021
|
|
5UP4
 
 | |
6IUK
 
 | | Cryo-EM structure of Murine Norovirus capsid | | Descriptor: | Major capsid protein VP1 | | Authors: | Song, C, Miyazaki, N, Iwasaki, K, Katayama, K, Murata, K. | | Deposit date: | 2018-11-28 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Dynamic rotation of the protruding domain enhances the infectivity of norovirus.
Plos Pathog., 16, 2020
|
|
6JI1
 
 | |
7NMZ
 
 | |
7ENB
 
 | |
7EMZ
 
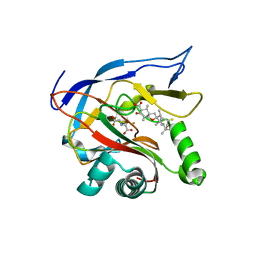 | |
7XWA
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Omicron BA.4/5 variant spike protein in complex with its receptor ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Suzuki, T, Kimura, K, Hashiguchi, T. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron BA.2 subvariants, including BA.4 and BA.5.
Cell, 185, 2022
|
|
5XPF
 
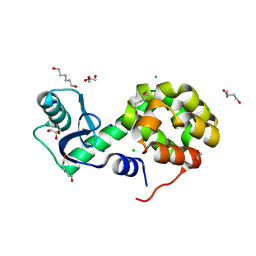 | | High-resolution X-ray structure of the T26H mutant of T4 lysozyme | | Descriptor: | CHLORIDE ION, Endolysin, GLYCEROL, ... | | Authors: | Hiromoto, T, Kuroki, R. | | Deposit date: | 2017-06-01 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Neutron structure of the T26H mutant of T4 phage lysozyme provides insight into the catalytic activity of the mutant enzyme and how it differs from that of wild type.
Protein Sci., 26, 2017
|
|
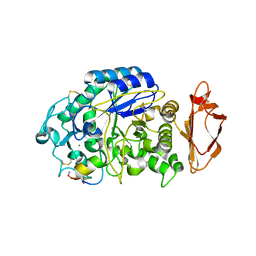
5VA9
 
 | |
5U3A
 
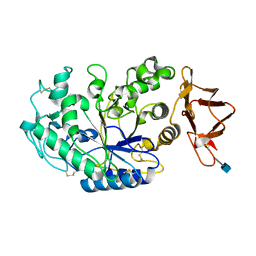 | |
5UV5
 
 | | Crystal Structure of a 2-Hydroxyisoquinoline-1,3-dione RNase H Active Site Inhibitor with Multiple Binding Modes to HIV Reverse Transcriptase | | Descriptor: | 7-(furan-2-yl)-2-hydroxyisoquinoline-1,3(2H,4H)-dione, MANGANESE (II) ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2017-02-19 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A 2-Hydroxyisoquinoline-1,3-Dione Active-Site RNase H Inhibitor Binds in Multiple Modes to HIV-1 Reverse Transcriptase.
Antimicrob. Agents Chemother., 61, 2017
|
|
5WZY
 
 | | Crystal structure of the P2X4 receptor from zebrafish in the presence of CTP at 2.8 Angstroms | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYTIDINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Kasuya, G, Hattori, M, Nureki, O. | | Deposit date: | 2017-01-19 | | Release date: | 2017-04-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Structural insights into the nucleotide base specificity of P2X receptors
Sci Rep, 7, 2017
|
|
5GW9
 
 | | Crystal structure of C163, a backbone circularized G-CSF | | Descriptor: | Granulocyte colony-stimulating factor | | Authors: | Miyafusa, T, Honda, S. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into the backbone-circularized granulocyte colony-stimulating factor containing a short connector.
Biochem. Biophys. Res. Commun., 500, 2018
|
|
5GMT
 
 | |
3LH0
 
 | |
3LM5
 
 | | Crystal Structure of human Serine/Threonine Kinase 17B (STK17B) in complex with Quercetin | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, Serine/threonine-protein kinase 17B | | Authors: | Ugochukwu, E, Soundararajan, M, Rellos, P, Fedorov, O, Phillips, C, Wang, J, Hapka, E, Filippakopoulos, P, Chaikuad, A, Pike, A.C.W, Carpenter, L, Vollmar, M, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-01-29 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A Chemical Probe for Dark Kinase STK17B Derives Its Potency and High Selectivity through a Unique P-Loop Conformation.
J.Med.Chem., 63, 2020
|
|
3LGF
 
 | |
3LGL
 
 | |
3RM1
 
 | |
2NLI
 
 | | Crystal Structure of the complex between L-lactate oxidase and a substrate analogue at 1.59 angstrom resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, HYDROGEN PEROXIDE, LACTIC ACID, ... | | Authors: | Furuichi, M, Suzuki, N, Balasundaresan, D, Yoshida, Y, Minagawa, H, Watanabe, Y, Kaneko, H, Waga, I, Kumar, P.K.R, Mizuno, H. | | Deposit date: | 2006-10-20 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | X-ray structures of Aerococcus viridans lactate oxidase and its complex with D-lactate at pH 4.5 show an alpha-hydroxyacid oxidation mechanism
J.Mol.Biol., 378, 2008
|
|
3RLZ
 
 | |
