8JNQ
 
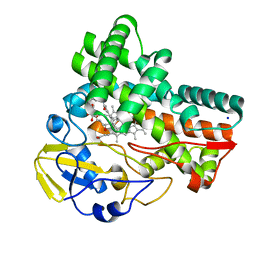 | | Crystal structure of cytochrome P450 CftA from Streptomyces torulosus NRRL B-3889, in complex with a substrate compound c | | Descriptor: | (1Z,3E,5E,7S,8R,10S,11R,13R,15R,16E,18E,25S)-11-ethyl-2,7-dihydroxy-10-methyl-21,26-diazatetracyclo[23.2.1.09,13.08,15]octacosa-1(2),3,5,16,18-pentaene-20,27,28-trione, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Jiang, P, Zhang, L.P, Zhang, C.S. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8JOO
 
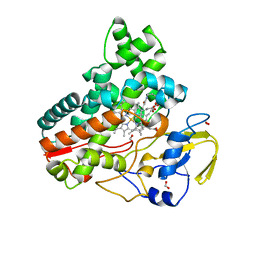 | | Crystal structure of cytochrome P450 IkaD from Streptomyces sp. ZJ306, in complex with the substrate ikarugamycin | | Descriptor: | (1Z,3E,5S,7R,8R,10R,11R,12S,15R,16S,18Z,25S)-11-ethyl-2-hydroxy-10-methyl-21,26-diazapentacyclo[23.2.1.05,16.07,15.08,12]octacosa-1(2),3,13,18-tetraene-20,27,28-trione, Cytochrome P450, FORMIC ACID, ... | | Authors: | Zhang, Y.L, Zhang, L.P, Zhang, C.S. | | Deposit date: | 2023-06-08 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8JUA
 
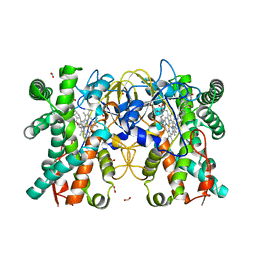 | | Multifunctional cytochrome P450 enzyme IkaD from Streptomyces sp. ZJ306, in complex with epoxyikarugamycin | | Descriptor: | (1Z,3E,5S,7R,8R,10R,11R,12S,13R,15S,16R,17S,19Z,26S)-11-ethyl-2-hydroxy-10-methyl-22,27-diaza-14 oxahexacyclo[24.2.1.05,17.07,16.013,15.08,12]nonacosa-1(2),3,19-triene-21,28,29-trione, Cytochrome P450, FORMIC ACID, ... | | Authors: | Zhang, Y.L, Zhang, L.P, Zhang, C.S. | | Deposit date: | 2023-06-26 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.00001121 Å) | | Cite: | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8JNO
 
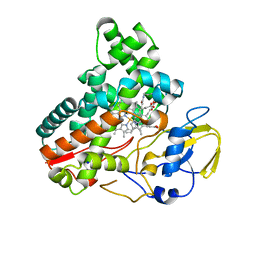 | | Crystal structure of cytochrome P450 IkaD from Streptomyces sp. ZJ306, in complex with the substrate 10-epi-deOH-HSAF | | Descriptor: | (1Z,3E,5S,7S,8R,9S,10S,11R,13R,15R,16S,18Z,25S)-11-ethyl-2,7-dihydroxy-10-methyl-21,26-diazapentacyclo[23.2.1.09,13.08,15.05,16]octacosa-1(2),3,18-triene-20,27,28-trione, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, Y.L, Zhang, L.P, Zhang, C.S. | | Deposit date: | 2023-06-06 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
4HGP
 
 | |
4JR6
 
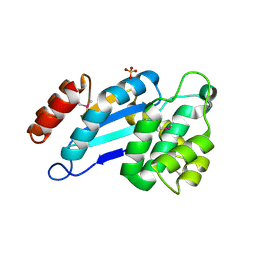 | | Crystal structure of DsbA from Mycobacterium tuberculosis (reduced) | | Descriptor: | Possible conserved membrane or secreted protein, SULFATE ION | | Authors: | Wang, L. | | Deposit date: | 2013-03-21 | | Release date: | 2013-07-17 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structure analysis of the extracellular domain reveals disulfide bond forming-protein properties of Mycobacterium tuberculosis Rv2969c.
Protein Cell, 4, 2013
|
|
2GUF
 
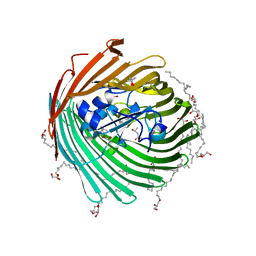 | | In meso crystal structure of the cobalamin transporter, BtuB | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FORMIC ACID, Vitamin B12 transporter btuB, ... | | Authors: | Caffrey, M, Cherezov, V, Yamashita, E, Cramer, W.A. | | Deposit date: | 2006-04-29 | | Release date: | 2006-12-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | In Meso Structure of the Cobalamin Transporter, BtuB, at 1.95 A Resolution.
J.Mol.Biol., 364, 2006
|
|
4JIA
 
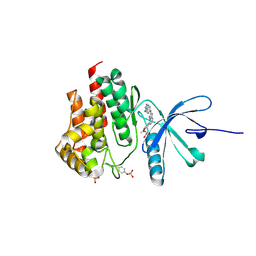 | | JAK2 kinase (JH1 domain) in complex with compound 9 | | Descriptor: | 5-(4-methoxyphenyl)-N-[4-(4-methylpiperazin-1-yl)phenyl][1,2,4]triazolo[1,5-a]pyridin-2-amine, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2013-03-05 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 2-Amino-[1,2,4]triazolo[1,5-a]pyridines as JAK2 inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4HGR
 
 | |
7YJ2
 
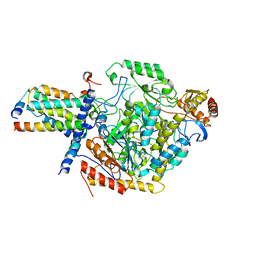 | | Cryo-EM structure of SPT-ORMDL3 (ORMDL3-N13A) complex | | Descriptor: | ORM1-like protein 3, PYRIDOXAL-5'-PHOSPHATE, Serine palmitoyltransferase 1, ... | | Authors: | Xie, T, Liu, P, Gong, X. | | Deposit date: | 2022-07-19 | | Release date: | 2023-07-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Ceramide sensing by human SPT-ORMDL complex for establishing sphingolipid homeostasis.
Nat Commun, 14, 2023
|
|
7YJ1
 
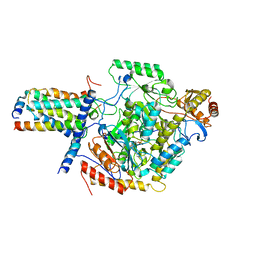 | | Cryo-EM structure of SPT-ORMDL3 (ORMDL3-deltaN2) complex | | Descriptor: | ORM1-like protein 3, PYRIDOXAL-5'-PHOSPHATE, Serine palmitoyltransferase 1, ... | | Authors: | Xie, T, Liu, P, Gong, X. | | Deposit date: | 2022-07-19 | | Release date: | 2023-07-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ceramide sensing by human SPT-ORMDL complex for establishing sphingolipid homeostasis.
Nat Commun, 14, 2023
|
|
4JI9
 
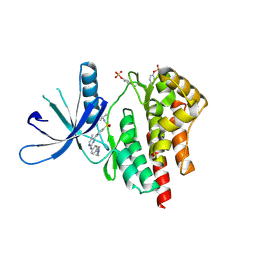 | | JAK2 kinase (JH1 domain) in complex with TG101209 | | Descriptor: | N-tert-butyl-3-[(5-methyl-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino]benzenesulfonamide, Tyrosine-protein kinase JAK2 | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2013-03-05 | | Release date: | 2013-08-07 | | Last modified: | 2013-08-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2-Amino-[1,2,4]triazolo[1,5-a]pyridines as JAK2 inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
7YIU
 
 | | Cryo-EM structure of the C6-ceramide-bound SPT-ORMDL3 complex | | Descriptor: | HEXADECANE, N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)HEXANAMIDE, ORM1-like protein 3, ... | | Authors: | Xie, T, Liu, P, Gong, X. | | Deposit date: | 2022-07-18 | | Release date: | 2023-07-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Ceramide sensing by human SPT-ORMDL complex for establishing sphingolipid homeostasis.
Nat Commun, 14, 2023
|
|
7YIY
 
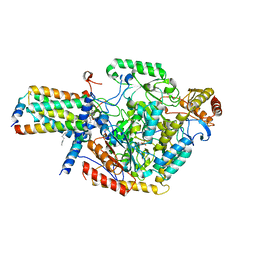 | | Cryo-EM structure of SPT-ORMDL3 complex | | Descriptor: | N-[(2S,3R,4E)-1,3-dihydroxyoctadec-4-en-2-yl]tetracosanamide, ORM1-like protein 3, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Xie, T, Liu, P, Gong, X. | | Deposit date: | 2022-07-18 | | Release date: | 2023-07-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Ceramide sensing by human SPT-ORMDL complex for establishing sphingolipid homeostasis.
Nat Commun, 14, 2023
|
|
4HGQ
 
 | |
4JR4
 
 | | Crystal structure of Mtb DsbA (Oxidized) | | Descriptor: | Possible conserved membrane or secreted protein, SULFATE ION | | Authors: | Wang, L. | | Deposit date: | 2013-03-21 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Structure analysis of the extracellular domain reveals disulfide bond forming-protein properties of Mycobacterium tuberculosis Rv2969c.
Protein Cell, 4, 2013
|
|
4HGN
 
 | |
4HGO
 
 | |
8FQ4
 
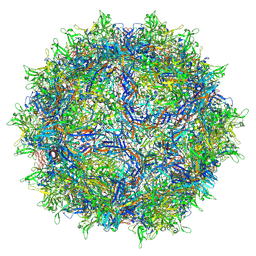 | | AAV1 VP3 Only Capsid | | Descriptor: | Capsid protein | | Authors: | Mietzsch, M, McKenna, R. | | Deposit date: | 2023-01-05 | | Release date: | 2023-02-08 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Production and characterization of an AAV1-VP3-only capsid: An analytical benchmark standard.
Mol Ther Methods Clin Dev, 29, 2023
|
|
8GPX
 
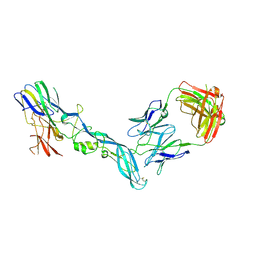 | | YFV_E_YD73Fab_postfusion | | Descriptor: | Envelope protein, YD73Fab_H, YD73Fab_K | | Authors: | Li, Y, Wu, L, Chai, Y, Qi, J, Yan, J, Gao, G.F. | | Deposit date: | 2022-08-27 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | A neutralizing-protective supersite of human monoclonal antibodies for yellow fever virus.
Innovation (N Y), 3, 2022
|
|
8GPU
 
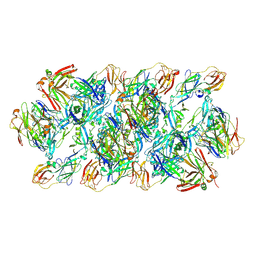 | | YFV_E_YD6Fab_prefusion | | Descriptor: | Envelope protein, YD6Fab_H, YD6Fab_L | | Authors: | Li, Y, Wu, L, Qi, J, Yan, J, Gao, G.F. | | Deposit date: | 2022-08-27 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | A neutralizing-protective supersite of human monoclonal antibodies for yellow fever virus.
Innovation (N Y), 3, 2022
|
|
8GPT
 
 | | YFV_E_YD6scFv_postfusion | | Descriptor: | Envelope protein, YD6_VH, YD6_VL | | Authors: | Li, Y, Wu, L, Qi, J, Yan, J, Gao, G.F. | | Deposit date: | 2022-08-27 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | A neutralizing-protective supersite of human monoclonal antibodies for yellow fever virus.
Innovation (N Y), 3, 2022
|
|
6AY5
 
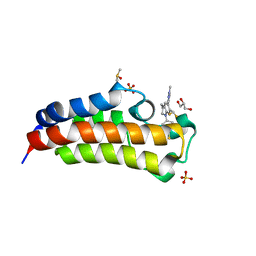 | | CREBBP bromodomain in complex with Cpd17 (5-(7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-3-methylbenzo[d]thiazol-2(3H)-one) | | Descriptor: | 5-[7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-3-methyl-1,3-benzothiazol-2(3H)-one, CREB-binding protein, DIMETHYL SULFOXIDE, ... | | Authors: | Murray, J.M. | | Deposit date: | 2017-09-07 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | A Unique Approach to Design Potent and Selective Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP) Inhibitors.
J. Med. Chem., 60, 2017
|
|
6AY3
 
 | | CREBBP bromodomain in complex with Cpd16 (5-(7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-N-methyl-1H-indole-3-carboxamide) | | Descriptor: | 1,2-ETHANEDIOL, 5-[7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-N-methyl-1H-indole-3-carboxamide, CREB-binding protein, ... | | Authors: | Murray, J.M. | | Deposit date: | 2017-09-07 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.391 Å) | | Cite: | A Unique Approach to Design Potent and Selective Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP) Inhibitors.
J. Med. Chem., 60, 2017
|
|
6C6P
 
 | | Human squalene epoxidase (SQLE, squalene monooxygenase) structure with FAD and NB-598 | | Descriptor: | (2E)-N-({3-[([3,3'-bithiophen]-5-yl)methoxy]phenyl}methyl)-N-ethyl-6,6-dimethylhept-2-en-4-yn-1-amine, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Padyana, A.K, Jin, L. | | Deposit date: | 2018-01-19 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and inhibition mechanism of the catalytic domain of human squalene epoxidase.
Nat Commun, 10, 2019
|
|
