3QUP
 
 | |
8QOE
 
 | | Inward-facing conformation of the ABC transporter BmrA | | Descriptor: | Multidrug resistance ABC transporter ATP-binding/permease protein BmrA | | Authors: | Di Cesare, M, Kaplan, E, Valimehr, S, Hanssen, E, Orelle, C, Jault, J.M. | | Deposit date: | 2023-09-28 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | The transport activity of the multidrug ABC transporter BmrA does not require a wide separation of the nucleotide-binding domains.
J.Biol.Chem., 300, 2023
|
|
3SN7
 
 | | Highly Potent, Selective, and Orally Active Phosphodiestarase 10A Inhibitors | | Descriptor: | 8-fluoro-6-methoxy-3,4-dimethyl-1-(3-methylpyridin-4-yl)imidazo[1,5-a]quinoxaline, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Parris, K.D. | | Deposit date: | 2011-06-28 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Highly Potent, Selective, and Orally Active Phosphodiesterase 10A Inhibitors.
J.Med.Chem., 54, 2011
|
|
5JO3
 
 | | PDE5A for NaV1.7 | | Descriptor: | 1-(5-chloro-6-methoxypyridin-3-yl)-3-methyl-N-(methylsulfonyl)-1H-indazole-5-carboxamide, ZINC ION, cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Storer, I, Chrencik, J. | | Deposit date: | 2016-05-01 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | PDE5A for NaV1.7
To be published
|
|
8RD2
 
 | |
3DV3
 
 | | MEK1 with PF-04622664 Bound | | Descriptor: | 3,4-difluoro-2-[(2-fluoro-4-iodophenyl)amino]-N-(2-hydroxyethoxy)-5-[(2-hydroxyethoxy)methyl]benzamide, ADENOSINE-5'-TRIPHOSPHATE, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Kazmirski, S.L, Kothe, M, Ding, Y.-H. | | Deposit date: | 2008-07-18 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Beyond the MEK-pocket: can current MEK kinase inhibitors be utilized to synthesize novel type III NCKIs? Does the MEK-pocket exist in kinases other than MEK?
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4FCB
 
 | | Potent and Selective Phosphodiesterase 10A Inhibitors | | Descriptor: | 3,4-dimethyl-1-propyl-7-(quinolin-2-ylmethoxy)imidazo[1,5-a]quinoxaline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Parris, K.D. | | Deposit date: | 2012-05-24 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel triazines as potent and selective phosphodiesterase 10A inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4FCD
 
 | | Potent and Selective Phosphodiesterase 10A Inhibitors | | Descriptor: | 1-(2-chlorophenyl)-6,8-dimethoxy-3-methylimidazo[5,1-c][1,2,4]benzotriazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Parris, K.D. | | Deposit date: | 2012-05-24 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Novel triazines as potent and selective phosphodiesterase 10A inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
7WGA
 
 | |
1RHD
 
 | |
3FQA
 
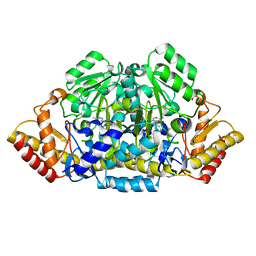 | | Gabaculien complex of gabaculine resistant GSAM version | | Descriptor: | 3-AMINOBENZOIC ACID, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Glutamate-1-semialdehyde 2,1-aminomutase | | Authors: | Stetefeld, J. | | Deposit date: | 2009-01-07 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Absence of a catalytic water confers resistance to the neurotoxin gabaculine.
Faseb J., 24, 2010
|
|
3FQ7
 
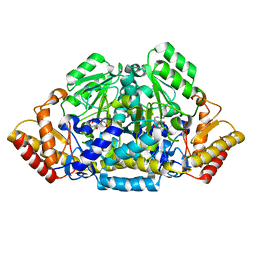 | | Gabaculine complex of GSAM | | Descriptor: | 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, Glutamate-1-semialdehyde 2,1-aminomutase | | Authors: | Stetefeld, J. | | Deposit date: | 2009-01-07 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Absence of a catalytic water confers resistance to the neurotoxin gabaculine.
Faseb J., 24, 2010
|
|
3FQ8
 
 | | M248I mutant of GSAM | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Glutamate-1-semialdehyde 2,1-aminomutase | | Authors: | Stetefeld, J. | | Deposit date: | 2009-01-07 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Absence of a catalytic water confers resistance to the neurotoxin gabaculine.
Faseb J., 24, 2010
|
|
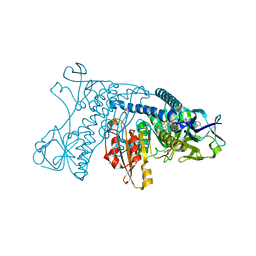
3GRS
 
 | |
3I84
 
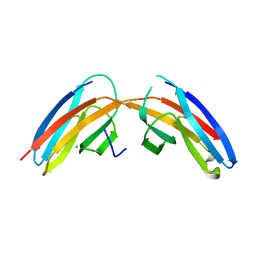 | |
3I85
 
 | |
3HS4
 
 | | Human carbonic anhydrase II complexed with acetazolamide | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Robbins, A.H, Genis, C, Domsic, J, Sippel, K.H, Agbandje-McKenna, M, McKenna, R. | | Deposit date: | 2009-06-10 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-resolution structure of human carbonic anhydrase II complexed with acetazolamide reveals insights into inhibitor drug design.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3AAG
 
 | | Crystal structure of C. jejuni pglb C-terminal domain | | Descriptor: | CALCIUM ION, General glycosylation pathway protein | | Authors: | Maita, N, Kohda, D. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Comparative structural biology of Eubacterial and Archaeal oligosaccharyltransferases.
J.Biol.Chem., 285, 2010
|
|
3V5Q
 
 | | Discovery of a selective TRK Inhibitor with efficacy in rodent cancer tumor models | | Descriptor: | 1-(3-{[(3Z)-2-oxo-3-(1H-pyrrol-2-ylmethylidene)-2,3-dihydro-1H-indol-6-yl]amino}phenyl)-3-[3-(trifluoromethyl)phenyl]urea, CHLORIDE ION, NT-3 growth factor receptor | | Authors: | Kreusch, A. | | Deposit date: | 2011-12-16 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2001 Å) | | Cite: | Discovery of GNF-5837, a Selective TRK Inhibitor with Efficacy in Rodent Cancer Tumor Models.
ACS Med Chem Lett, 3, 2012
|
|
4GRT
 
 | | HUMAN GLUTATHIONE REDUCTASE A34E, R37W MUTANT, MIXED DISULFIDE BETWEEN TRYPANOTHIONE AND THE ENZYME | | Descriptor: | BIS(GAMMA-GLUTAMYL-CYSTEINYL-GLYCINYL)SPERMIDINE, FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE | | Authors: | Stoll, V.S, Simpson, S.J, Krauth-Siegel, R.L, Walsh, C.T, Pai, E.F. | | Deposit date: | 1997-02-12 | | Release date: | 1997-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Glutathione reductase turned into trypanothione reductase: structural analysis of an engineered change in substrate specificity.
Biochemistry, 36, 1997
|
|
6ASP
 
 | | Structure of Grp94 with methyl 3-chloro-2-(2-(1-(2-ethoxybenzyl)-1 H-imidazol-2-yl)ethyl)-4,6-dihydroxybenzoate, a Grp94-selective inhibitor and promising therapeutic lead for treating myocilin-associated glaucoma | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Endoplasmin, GLYCEROL, ... | | Authors: | Huard, D.J.E, Lieberman, R.L. | | Deposit date: | 2017-08-25 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.696 Å) | | Cite: | Trifunctional High-Throughput Screen Identifies Promising Scaffold To Inhibit Grp94 and Treat Myocilin-Associated Glaucoma.
ACS Chem. Biol., 13, 2018
|
|
3WAK
 
 | |
6ASQ
 
 | | Structure of Grp94 bound to methyl 2-[2-(2-benzylpyridin-3-yl)ethyl]-3-chloro-4,6-dihydroxybenzoate, a pan-Hsp90 inhibitor | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, DI(HYDROXYETHYL)ETHER, Endoplasmin, ... | | Authors: | Huard, D.J.E, Lieberman, R.L. | | Deposit date: | 2017-08-25 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Trifunctional High-Throughput Screen Identifies Promising Scaffold To Inhibit Grp94 and Treat Myocilin-Associated Glaucoma.
ACS Chem. Biol., 13, 2018
|
|
6BI0
 
 | | Trastuzumab Fab N158A, D185A, K190A (Light Chain) Triple Mutant. | | Descriptor: | 1,2-ETHANEDIOL, Trastuzumab anti-HER2 Fab Heavy Chain, Trastuzumab anti-HER2 Fab Light Chain | | Authors: | DiDonato, M, Spraggon, G. | | Deposit date: | 2017-10-31 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.057 Å) | | Cite: | Tuning a Protein-Labeling Reaction to Achieve Highly Site Selective Lysine Conjugation.
Chembiochem, 19, 2018
|
|
6BI2
 
 | |
