6NZP
 
 | |
6NZQ
 
 | |
6NZH
 
 | |
8X91
 
 | | P/Q type calcium channel in complex with omega-conotoxin MVIIC | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Li, Z, Cong, Y, Wu, T, Wang, T. | | Deposit date: | 2023-11-29 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis for different omega-agatoxin IVA sensitivities of the P-type and Q-type Ca v 2.1 channels.
Cell Res., 34, 2024
|
|
8X90
 
 | | P/Q type calcium channel | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-[PHOSPHO-L-SERINE], 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Li, Z, Cong, Y, Wu, T, Wang, T. | | Deposit date: | 2023-11-29 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis for different omega-agatoxin IVA sensitivities of the P-type and Q-type Ca v 2.1 channels.
Cell Res., 34, 2024
|
|
8X93
 
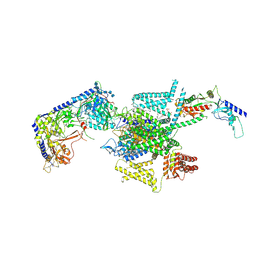 | | P/Q type calcium channel in complex with omega-Agatoxin IVA | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Li, Z, Cong, Y, Wu, T, Wang, T. | | Deposit date: | 2023-11-29 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structural basis for different omega-agatoxin IVA sensitivities of the P-type and Q-type Ca v 2.1 channels.
Cell Res., 34, 2024
|
|
4DAC
 
 | |
7BVC
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with ethambutol | | Descriptor: | 4'-PHOSPHOPANTETHEINE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BR3
 
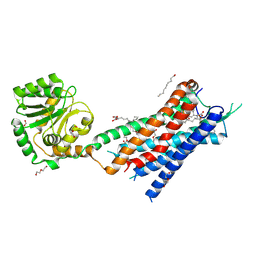 | | Crystal structure of the protein 1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R)-2,3-dihydroxypropyl dodecanoate, 4-[[(1R)-2-[5-(2-fluoranyl-3-methoxy-phenyl)-3-[[2-fluoranyl-6-(trifluoromethyl)phenyl]methyl]-4-methyl-2,6-bis(oxidanylidene)pyrimidin-1-yl]-1-phenyl-ethyl]amino]butanoic acid, ... | | Authors: | Cheng, L, Shao, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-10-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure of the human gonadotropin-releasing hormone receptor GnRH1R reveals an unusual ligand binding mode.
Nat Commun, 11, 2020
|
|
7BVE
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbC2-AcpM2 in complex with ethambutol | | Descriptor: | CALCIUM ION, Ethambutol, Integral membrane indolylacetylinositol arabinosyltransferase EmbC, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVF
 
 | | Cryo-EM structure of Mycobacterium tuberculosis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with ethambutol | | Descriptor: | CALCIUM ION, CARDIOLIPIN, Ethambutol, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BX8
 
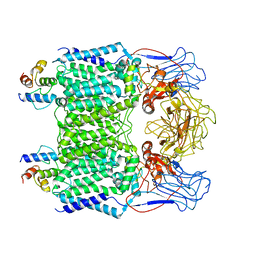 | | Mycobacterium smegmatis arabinosyltransferase complex EmbB2-AcpM2 in symmetric "resting state" | | Descriptor: | Integral membrane indolylacetylinositol arabinosyltransferase EmbB, Meromycolate extension acyl carrier protein | | Authors: | Gao, R.G, Zhang, L, Wang, Q, Rao, Z.H. | | Deposit date: | 2020-04-17 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM snapshots of mycobacterial arabinosyltransferase complex EmbB2-AcpM2.
Protein Cell, 11, 2020
|
|
6IZG
 
 | |
8JWY
 
 | | Crystal structure of A2AR-T4L in complex with 2-118 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[2-azanyl-6-[2-oxidanylidene-1-[[6-(2-oxidanylpropan-2-yl)pyridin-2-yl]methyl]pyridin-4-yl]pyrimidin-4-yl]-2-methyl-benzenecarbonitrile, Adenosine receptor A2a,Endolysin, ... | | Authors: | Weng, Y, Chen, Y, Xu, Y, Song, G. | | Deposit date: | 2023-06-29 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural insight into the dual-antagonistic mechanism of AB928 on adenosine A 2 receptors.
Sci China Life Sci, 67, 2024
|
|
8JWZ
 
 | | Crystal structure of A2AR-T4L in complex with AB928 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[2-azanyl-6-[1-[[6-(2-oxidanylpropan-2-yl)pyridin-2-yl]methyl]-1,2,3-triazol-4-yl]pyrimidin-4-yl]-2-methyl-benzenecarbonitrile, Adenosine receptor A2a,Endolysin, ... | | Authors: | Weng, Y, Chen, Y, Xu, Y, Song, G. | | Deposit date: | 2023-06-29 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural insight into the dual-antagonistic mechanism of AB928 on adenosine A 2 receptors.
Sci China Life Sci, 67, 2024
|
|
6IJA
 
 | | Crystal Structure of Arabidopsis thaliana UGT89C1 complexed with UDP-L-rhamnose | | Descriptor: | UDP-glycosyltransferase 89C1, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{R},4~{R},5~{R},6~{S})-6-methyl-3,4,5-tris(oxidanyl)oxan-2-yl] hydrogen phosphate | | Authors: | Zong, G, Wang, X. | | Deposit date: | 2018-10-09 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.214 Å) | | Cite: | Crystal structures of rhamnosyltransferase UGT89C1 from Arabidopsis thaliana reveal the molecular basis of sugar donor specificity for UDP-beta-l-rhamnose and rhamnosylation mechanism.
Plant J., 99, 2019
|
|
4NP4
 
 | | Clostridium difficile toxin B CROP domain in complex with FAB domains of neutralizing antibody bezlotoxumab | | Descriptor: | Toxin B, bezlotoxumab heavy chain, bezlotoxumab light chain | | Authors: | Orth, P, Xiao, L, Hernandez, L.D, Reichert, P, Sheth, P, Beaumont, M, Murgolo, N, Ermakov, G, DiNunzio, E, Racine, F, Karczewski, J, Secore, S, Ingram, R.N, Mayhood, T, Strickland, C, Therien, A.G. | | Deposit date: | 2013-11-20 | | Release date: | 2014-05-21 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Mechanism of Action and Epitopes of Clostridium difficile Toxin B-neutralizing Antibody Bezlotoxumab Revealed by X-ray Crystallography.
J.Biol.Chem., 289, 2014
|
|
4NJC
 
 | |
7BVH
 
 | | Crystal structure of arabinosyltransferase EmbC2-AcpM2 complex from Mycobacterium smegmatis complexed with di-arabinose | | Descriptor: | CALCIUM ION, Integral membrane indolylacetylinositol arabinosyltransferase EmbC, Meromycolate extension acyl carrier protein, ... | | Authors: | Zhao, Y, Zhang, L, Wu, L.J, Wang, Q, Li, J, Besra, G.S, Rao, Z.H. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
6IJD
 
 | | Crystal Structure of Arabidopsis thaliana UGT89C1 complexed with quercetin | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, UDP-glycosyltransferase 89C1 | | Authors: | Zong, G, Wang, X. | | Deposit date: | 2018-10-09 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.206 Å) | | Cite: | Crystal structures of rhamnosyltransferase UGT89C1 from Arabidopsis thaliana reveal the molecular basis of sugar donor specificity for UDP-beta-l-rhamnose and rhamnosylation mechanism.
Plant J., 99, 2019
|
|
8IF8
 
 | | Arabinosyltransferase AftA | | Descriptor: | CALCIUM ION, Galactan 5-O-arabinofuranosyltransferase | | Authors: | Gong, Y.C, Rao, Z.H, Zhang, L. | | Deposit date: | 2023-02-17 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the priming arabinosyltransferase AftA required for AG biosynthesis of Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3I6D
 
 | | Crystal structure of PPO from bacillus subtilis with AF | | Descriptor: | 5-[2-CHLORO-4-(TRIFLUOROMETHYL)PHENOXY]-2-NITROBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, ... | | Authors: | Shen, Y. | | Deposit date: | 2009-07-06 | | Release date: | 2009-12-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insight into unique properties of protoporphyrinogen oxidase from Bacillus subtilis
J.Struct.Biol., 170, 2010
|
|
7D6X
 
 | | Mycobacterium smegmatis Sdh1 complex in the apo form | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Zhou, X, Gao, Y, Wang, Q, Gong, H, Rao, Z. | | Deposit date: | 2020-10-02 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Architecture of the mycobacterial succinate dehydrogenase with a membrane-embedded Rieske FeS cluster.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7D6V
 
 | | Mycobacterium smegmatis Sdh1 in complex with UQ1 | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Zhou, X, Gao, Y, Wang, Q, Gong, H, Rao, Z. | | Deposit date: | 2020-10-02 | | Release date: | 2021-04-07 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Architecture of the mycobacterial succinate dehydrogenase with a membrane-embedded Rieske FeS cluster.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6IJ7
 
 | | Crystal Structure of Arabidopsis thaliana UGT89C1 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Rhamnosyltransferase protein | | Authors: | Zong, G, Wang, X. | | Deposit date: | 2018-10-09 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystal structures of rhamnosyltransferase UGT89C1 from Arabidopsis thaliana reveal the molecular basis of sugar donor specificity for UDP-beta-l-rhamnose and rhamnosylation mechanism.
Plant J., 99, 2019
|
|
